Abstract
Hyperspectral (HS) imaging (HSI) technology combines the main features of two existing technologies: imaging and spectroscopy. This allows to analyse simultaneously the morphological and chemical attributes of the objects captured by a HS camera. In recent years, the use of HSI provides valuable insights into the interaction between light and biological tissues, and makes it possible to detect patterns, cells, or biomarkers, thus, being able to identify diseases. This work presents the HistologyHSI-GB dataset, which contains 469 HS images from 13 patients diagnosed with brain tumours, specifically glioblastoma. The slides were stained with haematoxylin and eosin (H&E) and captured using a microscope at 20× power magnification. Skilled histopathologists diagnosed the slides and provided image-level annotations. The dataset was acquired using custom HSI instrumentation, consisting of a microscope equipped with an HS camera covering the spectral range from 400 to 1000 nm.
Similar content being viewed by others
Background & Summary
Hyperspectral (HS) imaging (HSI) is a technology able to measure both the spatial and spectral information of objects or substances, combining the features of spectroscopy and digital imaging in a single imaging modality. Because the absorption, reflection, transmission and scattering of light are unique to each material, this technology allows non-invasive identification of materials. The first use of HSI was for the remote sensing exploration of the Earth’s surface in the 80s1. In recent years, this technology has been extended to a wide range of applications, such as precision agriculture2,3, food quality inspection4,5,6, industrial sorting of materials7,8, art conservation9,10, or forensic sciences11,12. In medicine, recent research has proven HSI technology to be useful for different clinical applications13,14, for example, as a surgical guidance tool15,16, as a tool for early diagnosis17,18,19, or as a technology able to measure different biochemical parameters that can be useful for medical practitioners20,21,22,23.
Digital and computational pathology techniques are intended to provide pathologists with a tool for the quantitative analysis of pathological specimens, reducing inter-observer variability among different pathologists and saving the time of manual examination of histological specimens24,25. Recently, some researchers have investigated HSI as a suitable technology for computational pathology in various fields, such as digital staining, colour enhancement, standardization of pathological slides or the exploitation of autofluorescence or immunohistochemistry of histological slides26. However, the primary use of HSI in computational pathology is currently in diagnostic research for routine clinical practice. In this context, recent applications have been focused on the diagnosis of cholangiocarcinoma27,28, head and neck squamous cell carcinoma29, membranous nephropathy30, breast cancer31, or the classification of leukocyes32,33, among others.
The workflow in HS computational pathology research usually involves digitizing the histological slides using HSI instrumentation and extracting information from the HS images that could be useful for diagnostic purposes using various image processing methods. Although a wide variety of techniques are used in the literature to this end, it is difficult to compare the different approaches fairly, mainly due to the lack of publicly available datasets34.
In this work, we provide a publicly available dataset of HS images of haematoxylin and eosin (H&E) stained histological slides corresponding to brain tumours, specifically Glioblastoma (GB)35,36. To the best of our knowledge there are other databases related to gastric cancer but, this is the first publicly available dataset of HS brain histological images35. This dataset is composed of 469 HS images from 13 different patients, with image-level annotations for two different classes (non-tumour or tumour) according to the manual examination of the histological samples. The HS images cover the spectral range from 400 to 1000 nm and were taken at 20× magnification. On the one hand, this dataset can be relevant for researchers interested in HS image classification and other HS image processing techniques, such as spectral unmixing or HS data compression. This dataset was acquired by our research group and all HS images were employed to train different classification algorithms for GB detection which were presented in previous research work36,37,38. In this manuscript, we exclusively present the curated version of the dataset, from which artifacts and labelling errors found in previous publications have been eliminated. Besides the proposed classification techniques, a broad range of potential methods could be explored to evaluate the effectiveness of HSI in enhancing the performance relative to the outcomes obtained with RGB (Red-Green-Blue) images. On the other hand, this dataset can be used by researchers in the field of computational pathology and pathology practitioners to envision the possibilities of this technology for routine clinical practice. In this work, we provide a repository with the HS data, its homologous RGB image and, a snapshot of the original slides showing the region of interest for each HS image. We also provide a comprehensive explanation of the microscopic HS system, its quality validation process, and how the dataset is organized.
Methods
This section provides a detailed explanation of the methodology employed in previous works36,37,38, This includes a description of the methods used for collecting histological samples, an overview of the microscopic HS system, and the process of acquiring and processing the HS data.
Histological samples description
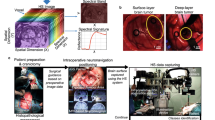
The research conducted in this study employs human biopsies extracted during brain tumour resection procedures (Fig. 1a). This research involved participants who were 18 years of age or older, all diagnosed with primary brain tumours and undergoing neurosurgical procedures at the University Hospital of Gran Canaria Doctor Negrín (Las Palmas de Gran Canaria, Spain). Prior to their involvement in the study, each participant provided written informed consent, which explicitly authorized the publication of any images or data obtained during the study. The Research Ethics Committee of the University Hospital of Gran Canaria Doctor Negrin (Comité Ético de Investigación Clínica-Comité de Ética en la Investigación, CEIC/CEI) approved the study protocol and consent procedures (reference 130069). All research procedures were conducted in strict compliance with applicable guidelines and regulations. The pathological slides used in this research were processed and analysed in the Pathological Anatomy Department of the same hospital. After the tumour tissue resection during neurosurgery, the biopsy samples underwent a series of standardized procedures. First, the samples were dehydrated to remove the excess of water, as it is immiscible with most embedding media. The samples were then embedded in paraffin blocks, mounted on microtomes and sliced into 4 µm thick slices. Finally, the slices were rehydrated and stained with H&E, a method commonly used in pathology.
The pathologists involved in the study analysed the stained sections using routine examination techniques. Each sample was evaluated and diagnosed as GB (a grade 4 primary brain tumour) according to the 2016 World Health Organization (WHO) classification of tumours of the central nervous system39. Macroscopic annotations of the GB locations on the physical pathological slides were made using a red marker pen (Fig. 1b). These annotations served as reference points for further analysis. In addition, non-tumour areas, where no discrete presence of tumour cells was observed, were annotated (blue marker pen) on the histology slides. The pen-marker annotations on the histological slide were deliberately outlined with wide borders to maintain a safety distance between tumour and non-tumour areas. Afterwards, regions of interest (ROIs) were selected from these pathologist-annotated areas for further study. These ROIs were subsequently digitized using the microscopic HS system (Fig. 1c), allowing for a detailed analysis of their spectral characteristics. Multiple HS images were acquired to cover the entire selected ROI. Figure 1d shows an example of the annotations within the pathological slide and the selection of different ROIs and the HS images (imaged at 20×). Finally, Fig. 1e summarizes the number of HS images acquired for each patient in the HistologyHSI-GB dataset.
Microscopic HS system
In this study, an HS camera coupled to a conventional brightfield microscope was employed to capture the HistologyHSI-GB dataset (Fig. 2). The HS camera (Fig. 2a) is a Hyperspec® VNIR A-Series from HeadWall Photonics (Fitchburg, MA, USA), which is based on an imaging spectrometer coupled to a CCD (charge-coupled device) sensor, the Adimec-1000 m (Adimec, Eindhoven, Netherlands). This HS camera works in the visual and near-infrared (VNIR) spectral range, from 400 to 1000 nm with a spectral resolution of 2.8 nm, sampling 1004 spatial pixels and, 826 spectral channels. The microscope is an Olympus BX-53 (Olympus, Tokyo, Japan), with four magnification lenses: 5×, 10×, 20× and 50×. The objective lenses are optimized for infrared (IR) observations and the light source is an halogen lamp (Fig. 2b). The HS camera is based on a push-broom technique, requiring a spatial scanning to acquire an HS cube. The system employs a mechanical stage (SCAN, Märzhäuser, Wetzlar, Germany) attached to the microscope for this purpose, which provides accurate movement in the 3 spatial axes directions (Fig. 2c-d). A more detailed description of the different parts of the acquisition system can be found in Table 1. A custom software was developed for synchronizing the scanning movement and the HS camera data acquisition. The optimal exposure time was configured to 40 ms (the maximum allowed by the HS camera). The scanning speed of the microscope platform was adjusted according to the ratio of pixel size to exposure time to obtain squared pixels in the resulting HS cubes. Since multiple images were captured from each ROI, the software was designed to enable the acquisition of consecutive HS cubes in a row. Whenever an HS cube was captured (composed by 800 lines), it was stored in memory while the camera and platform continuing to capture data until several cubes were captured. This approach helps save time during image acquisition and minimizes the need for human intervention. To prevent potential degradation of focus or errors caused by the platform while moving, the capture of consecutive HS cubes was limited to a maximum of ten.
Data acquisition methodology
As previously mentioned, relevant areas were identified on the slides and highlighted with a pen in blue (non-tumour) or red (tumour). The capture process of a sample starts by selecting a ROI, from different non-tumour and tumour highlighted areas, to be imaged. Since cells details are needed for further processing, a 20× magnification was chosen to capture the HS images. The coarse focus of the specimen (Fig. 3a) is performed using the microscope binoculars. The procedure relies on the user’s subjective criteria. The final HS image is brought into focus by examining a specific frame captured by the push-broom camera, referred to as the Yλ frame (e.g., Yλ frame extracted from the yellow line in Fig. 3b). The λ axis of an Yλ frame corresponds to the spectral information, while the Y axis represents the spatial information across the field of view (FOV) of the camera. The objective is to identify the sharpest spatial frequency along various working distances from the sensor to the sample (Fig. 3c shows a focused Yλ frame while Fig. 3d shows an unfocused one). The working distance adjustment is performed precisely by using the Z movement with the joystick.
Capture process to obtain focused HS cubes. (a) Example of a histology slide with tumour and non-tumour annotations. The yellow square identifies a ROI where the HS image was captured. (b) Synthetic RGB image where the Yλ frame employed to focus the sample is marked in yellow. Examples of (c) focused and (d) unfocused Yλ frames.
After achieving the optimal focus on the sample, the software was configured to capture several HS images consecutively, where the number of images is defined as an input parameter. The number of images should be kept relatively low to avoid the focus degradation throughout the specimen, due to the non-flat nature of microscopic samples and the platform error/vibration during movement. In this case, a maximum of 10 HS images were extracted consecutively from a selected ROI. The dataset was captured with the light power set to the maximum (100 W) and the exposure time to 40 ms. At 20× magnification, the pixel size is 0.373 µm, and the microscope platform was configured to scan the sample at a speed of 9.325 µm/s. Furthermore, to overcome the challenges posed by the high dimensionality of the HS images, the collected cubes were constrained to a spatial size of 800 lines, resulting in 1.23 GB data cubes. The HS cubes had a dimension of 800 × 1004 × 826 (number of lines × number of rows × number of bands), corresponding to a spatial size of 299 × 375 µm recorded over a span of 32 s.
After the HS images were captured, the reference images for calibration were acquired. In HS image processing, flat-field calibration is an essential step designed to correct the raw data recorded by an HS system. This method corrects the HS data for differences due to the environmental conditions and instrumentation. The flat-field calibration makes use of white (WR) and dark (DR) reference images. The WR recording is designed to capture data about the HS imaging system under the same conditions used for sample collection, without involving the sample itself. Therefore, the WR is obtained by scanning a section of the histological slide where no tissue is present. Since there is no sample material in such position of the slide, this HS frame contains the maximum values that the sensor is able to measure for each pixel and wavelength in the specified capturing conditions (exposure time, light intensity, the optical properties of the glass slide, etc.). Afterwards, the DR is captured by blocking the light transmission to the HS camera. This HS frame contains the minimum values that the system is able to provide for each pixel and band, and also information about the dark currents in the CCD. Ideally, the DR values should be very close to zero. However, higher values can be obtained, typically due to the intrinsic noise of the sensor. To ensure a robust measurement of the reference images for calibration, 100 Yλ frames are captured for both WR and DR, allowing any potential errors to be averaged. These reference images were employed for the HS data calibration as detailed in next section. During the acquisition process of the HistologyHSI-GB dataset, image-level annotations were applied. These annotations (tumour or non-tumour) remained consistent across the entire HS cube, indicating that all data within the cube shared the same annotation.
HS Data calibration
The goal of the HS microscopic system is to provide a spectral signature per spatial pixel of the captured scene. These spectral signatures indicate the percentage of incident radiation that the scanned object transmits or reflects at each captured wavelength. Various factors, including the inherent spectral response of the sensor, the transmission of light through lenses and optical components, and the spectral characteristics of the light source influence the spectral response of an HS acquisition system. To obtain spectral signatures that accurately indicate the percentage of transmitted or reflected radiation at each wavelength in the sample, the HS cubes need to be calibrated. This calibration consists of normalizing the captured HS pixels by linearly scaling their values considering the WR and DR. Equation (1) is employed to calibrate the HS data, where ri and Rawi refer to each Yλ frame from the calibrated and the raw image, respectively. Figure 4 shows an example of how the spectral signatures of different pixels (Fig. 4a) are scaled to transmittance using the aforementioned calibration. The shape of the WR and DR is shown in Fig. 4b, and several pixels from a ROI before (Fig. 4c), and after calibration (Fig. 4d).
Effect of calibration in the spectral signatures. (a) Grayscale image (generated by averaging each spectral band) and selecting pixels corresponding to different materials. (b) WR and DR spectral signatures. (c) Uncalibrated spectral signatures from the selected pixels. (d) Calibrated spectral signatures from the selected pixels. Colours in (c,d) correspond to selected pixels in (a).
Furthermore, the calibration process also helps to remove the stripping noise effect, which typically appears when acquiring HS images using push-broom scanners40. The stripping noise consists in spatially coherent lines that appear in the spatial scanning axis due to static artifacts produced in the sensors, which are repeated in each push-broom frame, as shown in Fig. 5a. In the calibrated images, the effect of the stripping noise disappears (Fig. 5b). The stripping noise is mainly produced due to the fact that different photo-receptors of the sensor have slightly different sensibilities, producing slightly different values when measuring exactly the same amount of incident radiation. The effect of stripping noise and light influence can also be observed in the synthetic RGB shown in Fig. 5c and how this effect disappears after performing the calibration (Fig. 5d).
Examples of the uncalibrated and calibrated HS images. (a,b) grayscale representation generated by averaging all spectral bands of the uncalibrated and calibrated HS images, respectively. (c,d) synthetic RGB image of the uncalibrated and calibrated HS images, respectively, generated using a model of human eye spectral response.
Greyscale images (Fig. 5a,b) were generated by averaging all spectral bands of the HS image, while the synthetic RGB image (Fig. 5c) was obtained closely mimicking the spectral response of the human eye41. For modelling the human eye spectral response, the method employed the normal probability density function following Eq. (2) over the HS data, where μ is the mean (μR = 590, μG = 560, and μB = 470) and σ is the standard deviation (σR = 0.08, σG = 0.06, and σB = 0.04). In Fig. 6, we can observe that, after the normal probability density function, RGB channels take the following central values and bandwidths: R = 590 ± 44 nm, G = 560 ± 79 nm and, B = 470 ± 111 nm.
Finally, in order to present some examples of the HistologyHSI-GB dataset, Fig. 7 shows the synthetic RGB images, as well as the different calibrated spectral bands found in several HS cubes. The contribution of the sensor noise can be observed in the extreme bands.
Data Records
The HistologyHSI-GB dataset42 has been deposited in The Cancer Imaging Archive (TCIA) repository43 for cancer imaging. The dataset is structured in a hierarchy of folders, as shown in Fig. 8. At the top level of the hierarchy there is a single folder associated with each one of the patients comprising the dataset. At the patient level, the folder names correspond to Pi, where \(\{i\in {\mathbb{N}}|1\le i\le 13\}\). For each patient, we can find several folders containing the different HS images for that patient. There is a different number of folders per patient, and the name of each folder encodes the information about which ROI of the histological slide the data was acquired from (ROI_j) and another field indicating an image identifier within that ROI (Ck). The folders in the image level also contain information about the image-level annotations according to the diagnosis, which can be tumour (T) or non-tumour (NT). The number of ROIs and image identifiers varies depending on the patient, but the total number of images from each class can be found in Fig. 1e. A conventional image of the slide with the macroscopic annotations and the location of the different ROIs within the slide is available for each patient (Pi.png).
Finally, each folder within the image-folder level contains an HS image from the histological slide, the necessary files for the calibration (dark and white references), and a synthetic RGB image extracted from the HS cube. The HS cubes are stored in ENVI format44 (the standard format for storing HS images). The ENVI format consists of a flat-binary raster file with an accompanying ASCII (American Standard Code for Information Interchange) header file. A more detailed description of the different files in each image folder can be found Table 2. The HS cubes from the histological slides and the white and dark references are stored as ENVI files. The HistologyHSI-GB dataset comprises 469 images from 13 different patients, where 166 images are labelled as tumour, and 303 are labelled as non-tumour.
Technical Validation
A technical validation was accomplished to support the quality of the HistologyHSI-GB dataset. Linear sensor systems demonstrate analogous basis functions for both spectral sensitivity and responsivity decomposition45. Spectral responsivity refers to the effectiveness of light detection in relation to its frequency or wavelength. However, camera channels often exhibit varying sensitivity across different wavelengths due to the spectral responsivities of the detectors and the non-uniform output of diffractive or filtering elements46. Proper characterization is essential for ensuring the reliability and accuracy of HS data analysis and interpretation. HS data captured for noise quantification and spectral and spatial calibration which are used to perform the technical validation (Fig. 9) can be found in a published dataset47.
Signal to noise ratio
In this section, we present the signal-to-noise ratio (SNR) measurements for our instrumentation. We obtained the signal (S) values by capturing images of the light without any sample, similar to the procedure used for recording the WR in flat-field calibration. For the noise (N) values, we recorded HS images in the absence of light. The SNR was calculated as the ratio between the mean value of S and the standard deviation of N. These recordings were taken over 100 push-broom frames under the same conditions as the image recordings. We calculated the SNR over the entire spectral range for the central pixel of the push-broom frame (Fig. 10a), which shows that the SNR exceeds 20 dB over the entire spectral range, peaking 42 dB at 655 nm. Furthermore, the SNR remains above 30 dB for wavelengths ranging from 448 to 894 nm. The SNR spatial distribution was also calculated over the camera FOV for different spectral bands (Fig. 10b), showing that SNR is evenly distributed over the FOV for the different spectral bands, indicating a uniform spatial distribution.
Spectral characterization
The WCT-2065 polymer (Fig. 9a), a transmittance wavelength calibration standard from Avian Technologies (New London, USA), was employed to conduct the spectral characterization of the microscopic HS system. It represents an alternative designation for NIST (National Institute of Standards and Technology) SRM-2065 standard48. Its purpose lies in facilitating the calibration of spectrophotometers, covering the wavelength range of 400–2200 nm. The standard uses a glass filter material that incorporates a combination of rare earth oxides. This glass composition includes holmium oxide, samarium oxide, ytterbium oxide, and neodymium oxide, which are blended with lanthanum, boron, silicon, and zirconium oxides found in the base glass. The resulting combination of these oxides creates a filter material with specific optical properties suitable for calibration purposes.
An HS image of the WCT-2065 polymer was captured using the microscopic HS system and further pre-processed. This calibration standard can qualitatively validate the spectral quality of the employed instrumentation (Fig. 11). However, a systematic approach is required for a more accurate and thorough calibration. In order to perform the quantitative validation, the Pearson correlation coefficient (PCC) and root mean square error (RMSE) Eq. (3) were employed to measure the difference between two sets of data. PCC measure the degree of linear anti-correlation or correlation in the range [−1, 1], where −1 indicates perfectly linearly anti-correlated data and 1 indicates perfectly linearly correlated data and it is computed following the Eq. (4). In addition, local maxima and minima were found to detect the most significant signal peaks. Thus, similar peaks were identified both in the captured image and the reference (represented by red and black crosses in Fig. 11a, respectively), resulting in a mean wavelength difference spectra shift of 6.60 nm between them. Furthermore, Fig. 11b shows that the PCC between WCT-2065 and the measured HS peak absorbance values provided good value, as well as, it RMSE (PCC = 1 and RMSE = 0.02). Thus, this NIST traceable standard allows accurate and reliable measurements of the spectral reliability of the HS acquisition system.
Spatial characterization
Spatial resolution, the ability of a camera to capture fine details and distinguish between separate objects, is also a critical feature in imaging systems. It determines the smallest size of an object that can be recorded. This parameter is essential in applications like histological diagnosis, where identifying small details is essential. Accurate spatial resolution characterization enables improved system performance and precise analysis in various fields, including histopathology49. Firstly, the camera is manually aligned to capture the information properly50. Then, the spatial resolution of the microscopic HS system was evaluated both theoretically and empirically. The theoretical calculation of the FOV, shown in Eq. (5), considered factors such as pixel size (Ps), number of pixels (N), magnification (Mi), and sensor size (Ss).
An empirical test using a micrometre ruler (Fig. 9b) provides further insight into the spatial resolution capabilities of the cameras. In order to perform this test, a Yλ spatial profile of the ruler (Fig. 12a) and its first derivative (Fig. 12b) was analysed to determine the mean distance between peaks (local minima peaks signalled with red crosses and local maxima peaks with green crosses). The results of the theoretical calculation provide a pixel size of 0.3700 μm and the empirical one is 0.3697 μm. This method confirmed that the spatial resolution of the microscopic HS system matches the theoretical pixel size with an average error of less than 0.0003 μm.
Once the pixel size has been calculated and the camera has been visually aligned, together with the information about the mechanical resolution and frame rate of the camera, the required motor rotation speed of the mechanical stage is determined. However, an additional analysis was conducted to further improve and verify the correct configuration of the scanning parameters. The entire HS acquisition system is considered as a whole, including the microscope, camera, and movement mechanism. For this evaluation, the goal is to capture an image of a circle printed in a calibration slide (dot target) and evaluate its appearance to identify camera misalignments and suboptimal movement speeds. The circle appears as a perfect rounded circle when captured at the correct speed but appears as an ellipse when the speed is too high or too low. While visual inspection provides a relatively good assessment, an automatic methodology50 is needed for a more precise and rigorous calibration. First, principal component analysis (PCA) is employed to find the directions of the longest and shortest axes of the ellipse (ϕmin and ϕmax). Then, eccentricity can be calculated following Eq. (6), where a perfect circle would provide values close to zero. In our case, the dot target from the calibration slide was captured, and its eccentricity was computed providing accurate results (e = 0.176). Thus, the microscopic HS system is properly calibrated in the spatial domain, and it is possible to acquire HS images under satisfactory conditions.
Usage Notes
Recommended pre-processing
The pre-processing framework applied to each HS cube is based on standard calibration and spectral band reduction. First, HS images are transformed from radiance to normalized transmittance by calibration. As a result of the strong correlation of spectral information between adjacent spectral bands, we propose to reduce the spectral dimensionality of the original data. A spectrally reduced HS image is generated by averaging the spectral bands of adjacent neighbouring bands to perform this band reduction. Using a spectral window of three neighbours, this process reduces the original 826 bands to 275, while slightly decreasing the presence of white Gaussian noise. Furthermore, reducing the number of bands proves to be advantageous in terms of reducing the computational cost of subsequent image processing tasks. However, this band reduction is optional, depending on the further processing interest. Additionally, for image analysis involving the spectral analysis of the samples, it is recommended to perform a background sample segmentation, where the pixels corresponding to the tissue and the background light of the microscope are identified. Finally, to use classification methods, the label (tumour or non-tumour) of each HS cube should be extracted from the folder name.
Recommended data partition and data HS processing applications
To perform machine learning analysis, an unbiased data partition should be performed. The dataset used for this study poses three challenges. First, the dataset is limited in the number of patients (13 patients). Second, samples containing both classes (tumour and non-tumour) are only available for 8 patients. Hence, the non-tumour samples information is limited in terms of patients. Third, the dataset is unbalanced, with more images annotated as non-tumour. In previous works36,38, a data partition based on 4 different folds was employed. Furthermore, spectral unmixing techniques could be performed as a preprocessing stage prior to classification51, or they can be used to determine the abundances of known endmembers of the images, specifically identifying the proportions of the H&E stains in each pixel52.
Limitations and future perspectives
The dataset has several limitations. As previously mentioned, the primary limitation is its relatively small cohort, consisting of data from only 13 patients. Furthermore, information for both classes of interest, tumour, and non-tumour, is available for only 8 of these patients. This leads to an imbalanced dataset, with a predominance of images classified as non-tumour. Such an imbalance could potentially introduce bias and affect the generalizability of the findings derived from this dataset.
Another limitation is related to the type of annotations available in this dataset. The macroscopic annotations of tumour and non-tumour regions on the pathological slides, leading to only image-level annotations for the HS images. A more sophisticated method for digitally annotating the images would allow to identify regions where tumour and non-tumour tissues are adjacent, making possible to capture regions comprising both classes in a single HS image. More detailed digital annotation would help in further validating the classification algorithms on a pixel-by-pixel basis and could also offer potential for other methods such as unsupervised learning or spectral unmixing. However, more detailed annotations would significantly increase the time and manual effort required to label each image.
Finally, this dataset is focused on images captured on a single magnification (20×). The motivation of using the higher magnification available for the instrumentation was driven by the need to capture detailed cell-level information from the histological slides. However, creating a dataset containing the same images at different magnifications could be of potential interest and benefit to the scientific community.
In summary, future datasets of HS histological samples will need to include a larger number of patients, ensure a balanced representation of the various classes of interest, incorporate more detailed annotations, and provide images at various magnification levels.
Code availability
A tutorial on how to read and display HS data is available in a public repository: https://github.com/HIRIS-Lab/HistologyHSI-GB. These tutorials include the use of custom MATLAB and Python functions and some of the most common toolbox/libraries.
References
Goetz, A. F. H., Vane, G., Solomon, J. E. & Rock, B. N. Imaging Spectrometry for Earth Remote Sensing. Science (1979) 228, 1147–1153 (1985).
Khan, A., Vibhute, A. D., Mali, S. & Patil, C. H. A systematic review on hyperspectral imaging technology with a machine and deep learning methodology for agricultural applications. Ecol Inform 69, 101678 (2022).
Sethy, P. K., Pandey, C., Sahu, Y. K. & Behera, S. K. Hyperspectral imagery applications for precision agriculture - a systemic survey. Multimed Tools Appl 81, 3005–3038 (2022).
Saha, D. & Manickavasagan, A. Machine learning techniques for analysis of hyperspectral images to determine quality of food products: A review. Curr Res Food Sci 4, 28–44 (2021).
Ortega, S., Lindberg, S.-K., E. Anderssen, K. & Heia, K. Perspective Chapter: Hyperspectral Imaging for the Analysis of Seafood. in Hyperspectral Imaging - A Perspective on Recent Advances and Applications https://doi.org/10.5772/intechopen.108726 (IntechOpen, 2023).
Kang, Z. et al. Advances in Machine Learning and Hyperspectral Imaging in the Food Supply Chain. Food Engineering Reviews 14, 596–616 (2022).
Zheng, Y., Bai, J., Xu, J., Li, X. & Zhang, Y. A discrimination model in waste plastics sorting using NIR hyperspectral imaging system. Waste Management 72, 87–98 (2018).
Bonifazi, G., Capobianco, G. & Serranti, S. A hierarchical classification approach for recognition of low-density (LDPE) and high-density polyethylene (HDPE) in mixed plastic waste based on short-wave infrared (SWIR) hyperspectral imaging. Spectrochim Acta A Mol Biomol Spectrosc 198, 115–122 (2018).
Cucci, C. et al. Reflectance hyperspectral data processing on a set of Picasso paintings: which algorithm provides what? A comparative analysis of multivariate, statistical and artificial intelligence methods. in Optics for Arts, Architecture, and Archaeology VIII (eds. Groves, R. & Liang, H.) 1. https://doi.org/10.1117/12.2593838 (SPIE, 2021).
Balas, C., Epitropou, G., Tsapras, A. & Hadjinicolaou, N. Hyperspectral imaging and spectral classification for pigment identification and mapping in paintings by El Greco and his workshop. Multimed Tools Appl 77, 9737–9751 (2018).
Huang, S.-Y. et al. Recent Advances in Counterfeit Art, Document, Photo, Hologram, and Currency Detection Using Hyperspectral Imaging. Sensors 22, 7308 (2022).
Koz, A. Ground-Based Hyperspectral Image Surveillance Systems for Explosive Detection: Part I—State of the Art and Challenges. IEEE J Sel Top Appl Earth Obs Remote Sens 12, 4746–4753 (2019).
Karim, S., Qadir, A., Farooq, U., Shakir, M. & Laghari, A. A. Hyperspectral Imaging: A Review and Trends towards Medical Imaging. Curr Med Imaging Rev 19, 417–427 (2022).
Fei, B. Hyperspectral imaging in medical applications. in 523–565. https://doi.org/10.1016/B978-0-444-63977-6.00021-3 (2019).
Barberio, M. et al. Intraoperative Guidance Using Hyperspectral Imaging: A Review for Surgeons. Diagnostics 11, 2066 (2021).
Shapey, J. et al. Intraoperative multispectral and hyperspectral label‐free imaging: A systematic review of in vivo clinical studies. J Biophotonics 12 (2019).
Mangotra, H., Srivastava, S., Jaiswal, G., Rani, R. & Sharma, A. Hyperspectral imaging for early diagnosis of diseases: A review. Expert Syst https://doi.org/10.1111/exsy.13311 (2023).
Johansen, T. H. et al. Recent advances in hyperspectral imaging for melanoma detection. WIREs Computational Statistics 12 (2020).
Reshef, E. R., Miller, J. B. & Vavvas, D. G. Hyperspectral Imaging of the Retina: A Review. Int Ophthalmol Clin 60, 85–96 (2020).
Fouad Aref, M. H., Sharawi, A. A. R. & El-Sharkawy, Y. H. Delineation of the Arm Blood Vessels Utilizing Hyperspectral Imaging to Assist with Phlebotomy for Exploiting the Cutaneous Tissue Oxygen Concentration. Photodiagnosis Photodyn Ther 33, 102190 (2021).
Tomanic, T. et al. Estimating quantitative physiological and morphological tissue parameters of murine tumor models using hyperspectral imaging and optical profilometry. J Biophotonics 16 (2023).
Köhler, H. et al. Evaluation of hyperspectral imaging (HSI) for the measurement of ischemic conditioning effects of the gastric conduit during esophagectomy. Surg Endosc 33, 3775–3782 (2019).
Sucher, R. et al. Hyperspectral Imaging (HSI) of Human Kidney Allografts. Ann Surg 276, e48–e55 (2022).
Fuchs, T. J. & Buhmann, J. M. Computational pathology: Challenges and promises for tissue analysis. Computerized Medical Imaging and Graphics 35, 515–530 (2011).
Louis, D. N. et al. Computational Pathology: A Path Ahead. Arch Pathol Lab Med 140, 41–50 (2016).
Ortega, S., Halicek, M., Fabelo, H., Callico, G. M. & Fei, B. Hyperspectral and multispectral imaging in digital and computational pathology: a systematic review [Invited. Biomed Opt Express 11, 3195 (2020).
Sun, L. et al. Diagnosis of cholangiocarcinoma from microscopic hyperspectral pathological dataset by deep convolution neural networks. Methods 202, 22–30 (2022).
Deng, Y. et al. ResNet-50 based Method for Cholangiocarcinoma Identification from Microscopic Hyperspectral Pathology Images. J Phys Conf Ser 1880, 012019 (2021).
Ma, L. et al. Automatic detection of head and neck squamous cell carcinoma on histologic slides using hyperspectral microscopic imaging. J Biomed Opt 27 (2022).
Lv, M. et al. Membranous nephropathy classification using microscopic hyperspectral imaging and tensor patch-based discriminative linear regression. Biomed Opt Express 12, 2968 (2021).
Wang, J. et al. PCA-U-Net based breast cancer nest segmentation from microarray hyperspectral images. Fundamental Research 1, 631–640 (2021).
Duan, Y. et al. Leukocyte classification based on spatial and spectral features of microscopic hyperspectral images. Opt Laser Technol 112, 530–538 (2019).
Wang, Q. et al. A 3D attention networks for classification of white blood cells from microscopy hyperspectral images. Opt Laser Technol 139, 106931 (2021).
Ortega, S. et al. Information Extraction Techniques in Hyperspectral Imaging Biomedical Applications. in Multimedia Information Retrieval. https://doi.org/10.5772/intechopen.93960 (IntechOpen, 2021).
Zhang, Y., Wang, Y., Zhang, B. & Li, Q. A hyperspectral dataset of precancerous lesions in gastric cancer and benchmarks for pathological diagnosis. J Biophotonics 15 (2022).
Ortega, S. et al. Hyperspectral Imaging for the Detection of Glioblastoma Tumor Cells in H&E Slides Using Convolutional Neural Networks. Sensors 20, 1911 (2020).
Ortega Sarmiento, S. Automatic classification of histological hyperspectral images: algorithms and instrumentation, Universidad de Las Palmas de Gran Canaria. https://hdl.handle.net/10553/107311 (2021).
Ortega, S. et al. Hyperspectral Superpixel-Wise Glioblastoma Tumor Detection in Histological Samples. Applied Sciences 10, 4448 (2020).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathologica vol. 131 803–820 (Springer Berlin Heidelberg, 2016).
Gómez-Chova, L. et al. Correction of systematic spatial noise in push-broom hyperspectral sensors: application to CHRIS/PROBA images. Appl Opt 47, F46–60 (2008).
Wald, G. The Receptors of Human Color Vision. Science (1979) 145, 1007–1016 (1964).
Ortega, S. et al. Hyperspectral Histological Images for Diagnosis of Human Glioblastoma (HistologyHSI-GB). https://doi.org/10.7937/Z1K6-VD17 (2024).
Clark, K. et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J Digit Imaging 26, 1045–1057 (2013).
Triglav, J. Exelis Visual Information Solutions. Geoinformatics, Emmeloord 15, 34–37 (2012).
Physics-Based Vision: Principles and Practice. https://doi.org/10.1201/9781439865880 (A K Peters/CRC Press, 1993).
Pekkala, O., Pulli, T., Kokka, A. & Ikonen, E. Setup for characterising the spectral responsivity of Fabry–Pérot-interferometer-based hyperspectral cameras. Metrologia 56, 065005 (2019).
Ortega, S. et al. Technical Validation data to support the quality of the HistologyHSI-GB dataset. Figshare, https://doi.org/10.6084/m9.figshare.23659170.v1 (2023).
Choquette, S. J., Duewer, D. L., Hanssen, L. M. & Early, E. A. Standard Reference Material 2036 Near-Infrared Reflection Wavelength Standard. Appl Spectrosc 59, 496–504 (2005).
Intriligator, J. & Cavanagh, P. The Spatial Resolution of Visual Attention. Cogn Psychol 43, 171–216 (2001).
Ortega, S. et al. Hyperspectral Push-Broom Microscope Development and Characterization. IEEE Access 7, 122473–122491 (2019).
Cruz-Guerrero, I. A. et al. Hybrid Brain Tumor Classification Scheme of Histopathology Hyperspectral Images Using Linear Unmixing and Deep Learning. SSRN Electronic Journal https://doi.org/10.2139/ssrn.4292742 (2022).
Gibbs, S. L. et al. Near-Infrared Fluorescent Digital Pathology for the Automation of Disease Diagnosis and Biomarker Assessment. Mol Imaging 14, 7290.2015.00005 (2015).
Acknowledgements
This work has been supported by Spanish Government and European Union (FEDER funds) in the context of TALENT-HExPERIA (HypErsPEctRal Imaging for Artificial intelligence applications) project (PID2020-116417RB-C42 AEI/10.13039/501100011033). Moreover, this work was completed while Laura Quintana-Quintana and Raquel Leon were beneficiary of the pre-doctoral grant given by the “Agencia Canaria de Investigacion, Innovacion y Sociedad de la Información (ACIISI)” of the “Consejería de Economía, Conocimiento y Empleo”, which is part-financed by the European Social Fund (FSE) (POC 2014–2020, Eje 3 Tema Prioritario 74 (85%)). Furthermore, Himar Fabelo was also beneficiary of the FJC2020-043474-I funded by MCIN/AEI/10.13039/501100011033 and by the European Union “NextGenerationEU/PRTR”.
Author information
Authors and Affiliations
Contributions
S.O. designed and established the experimental system, conceived the study, generated the hyperspectral dataset, performed image processing and result analysis, and wrote the manuscript. L.Q.-Q. deposited the dataset, conducted the technical validation experiments, performed image processing and result analysis, and wrote the manuscript. R.L. deposited the dataset, supervised the technical validation, performed image processing and result analysis, and wrote the manuscript. H.F. Generated the hyperspectral dataset, supervised the study, and reviewed the manuscript. M.P. and R.C. prepared and diagnosed the histological samples and reviewed the manuscript. G.M.C. acquired the funding, supervised the study, and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ortega, S., Quintana-Quintana, L., Leon, R. et al. Histological Hyperspectral Glioblastoma Dataset (HistologyHSI-GB). Sci Data 11, 681 (2024). https://doi.org/10.1038/s41597-024-03510-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03510-x
- Springer Nature Limited