Abstract
Advancements in dermatological artificial intelligence research require high-quality and comprehensive datasets that mirror real-world clinical scenarios. We introduce a collection of 18,946 dermoscopic images spanning from 2010 to 2016, collated at the Hospital Clínic in Barcelona, Spain. The BCN20000 dataset aims to address the problem of unconstrained classification of dermoscopic images of skin cancer, including lesions in hard-to-diagnose locations such as those found in nails and mucosa, large lesions which do not fit in the aperture of the dermoscopy device, and hypo-pigmented lesions. Our dataset covers eight key diagnostic categories in dermoscopy, providing a diverse range of lesions for artificial intelligence model training. Furthermore, a ninth out-of-distribution (OOD) class is also present on the test set, comprised of lesions which could not be distinctively classified as any of the others. By providing a comprehensive collection of varied images, BCN20000 helps bridge the gap between the training data for machine learning models and the day-to-day practice of medical practitioners. Additionally, we present a set of baseline classifiers based on state-of-the-art neural networks, which can be extended by other researchers for further experimentation.
Similar content being viewed by others
Background & Summary
Skin cancer is one of the most frequent types of cancer and manifests mainly in areas of the skin most exposed to the sun. Despite not being the most frequent of the skin cancers, melanoma is responsible for 75% of deaths from skin tumours1. It can appear at any age, and it is the first most diagnosed cancer among patients in the 25–29 age group, the second among 20–24 years old, and the third among 15–19 years old2. Melanoma leads to significant years of productive life lost and stands as the costliest skin cancer in Europe in terms of cost per death3.
Given that skin cancer lesions are visible in the skin, dermatoscopy is a crucial tool in the diagnosis of skin cancer. This technique employs a magnifying lens and polarized light, penetrating deeper within the skin layers and minimizing surface reflection. Prior research has found that this technique allows enhanced visualization of the lesion structures, improving dermatologists’ diagnostic accuracy4,5. While experts typically look for specific structural and colour cues, standardized clinical procedures, such as the “ABCD rule of dermoscopy”, have been instrumental in skin cancer diagnosis6.
With the increasing use of high-resolution cameras and specialized dermoscopic adapters in medical institutions, there has been a surge in dermoscopic image data, prompting the development of computer vision algorithms for automatic lesion diagnosis7,8,9.
Earlier systems relied on the extraction of handcrafted features from skin lesions, similar to the rule sets that clinicians were using to perform diagnosis10,11,12,13. The aim was to develop specialized algorithms extracting colour, border features, symmetry, and other diagnostic criteria as inputs for machine learning classifiers. However, the growing availability of dermoscopic images has led to more sophisticated deep learning algorithms, particularly convolutional neural networks, that process images directly without relying on predefined rule sets. A significant catalyst for these algorithms’ adoption has been The International Skin Imaging Collaboration (ISIC), organizing annual challenges since 2016 for developing computer vision algorithms to classify and segment dermoscopic images of skin lesions14,15,16,17,18. Tschandl et al. showed that the performance of expert dermatologists confronted with dermoscopic images was outperformed by the top-scoring machine learning classifiers of the ISIC 2018 Challenge16,19. However, as highlighted by the authors, these algorithms underperformed on out-of-distribution images not represented in the training dataset of HAM1000020. A meta-study21 on the ISIC 2019 challenge17 has shown that state-of-the-art classification methods decrease by more than 20% on datasets specifically designed to better reflect clinical realities, as compared with a previous, well-controlled benchmark.
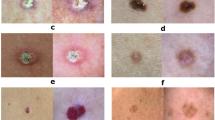
The Hospital Clinic of Barcelona is a tertiary referral center. Its department of dermatology is responsible for treating high-risk melanoma patients. The hospital often receives challenging cases from other regional centers, making it a representative sample of lesions encountered across the region. The BCN20000 dataset addresses the challenge of unconstrained classification of dermoscopic images of skin cancer, capturing a wide array of lesions in diagnostically challenging locations (nails and mucosa), alongside not segmentable and hypopigmented lesions. Figure 1 showcases some of these diagnostic challenges. Most importantly, this dataset, aptly termed ‘dermoscopic lesions in the wild’, contains OOD images which occur in normal clinical practice but are not represented in current dermoscopic datasets.
Comprising 18946 dermoscopic images corresponding to 5583 skin lesions, the dataset includes nevus, melanoma, basal cell carcinoma, seborrheic keratosis, actinic keratosis, squamous cell carcinoma, dermatofibroma, vascular lesion and OOD lesions that do not fit into the other categories. Figure 2 illustrates an example of the training categories.
Together with the images, we provide information related to the anatomic location of the lesion, age, capture date, and sex of the patients. Our efforts were directed to create a diverse dataset with cases that dermatologist face in their usual clinical practice. We also present the results obtained after training six baseline algorithms to classify images into the eight lesion categories.
Methods
General
The BCN20000 dataset comprises 18946 high-quality dermoscopic images corresponding to 5583 skin lesions captured from 2010 to 2016. These images were collected by the Department of Dermatology at the “Hospital ClÃnic de Barcelona”, employing a set of dermoscopic attachments on three high-resolution cameras. Images were originally stored in a secure server using a directory structure organized by camera and capture month. Informed consent was obtained from all participants.
The dataset is published open source for research, to enable the training of classification algorithms such as artificial neural networks. The dataset is split into train and test sets, with 12413 and 6533 images, respectively. Diagnosis labels are only provided for the training set. The separation of the dataset into training and test sets is due to its inclusion in the broader ISIC database. Establishing a private test set is crucial for the challenges organized by the ISIC consortium16,18,22, allowing for unbiased evaluation of algorithmic performance. Specifically, all images classified as ‘unknown’ were assigned to the test set. For the remaining images, we employed stratified random sampling to create the training set.
Image acquisition
Every time a new patient came to the hospital, the doctors captured a photo of the identifier sticker containing the name and unique patient identifier. Then, the doctors captured several clinical and close-up pictures, and finally, they obtained one or more dermoscopic images, see Fig. 3. The total number of images captured from 2010 until 2016 is 94302. These images were organized in folders per camera and month of capture, without differentiation of the imaging types (sticker, clinical, close-up or dermatoscopy images). Images were collected and shared with institutional ethics approval number HCB/2019/0413.
BCN20000 Dataset Preparation Pipeline. The process begins with the collection of images and metadata. A neural network is then employed to classify and separate between the image types. Patient identifiers are extracted from ‘Sticker pictures’ using a YOLOv3 network. Dermatoscopy’s diagnosis are revised by multiple reviewers for quality assurance. The resultant BCN20000 dataset is composed exclusively of dermoscopic images and metadata, divided into training and testing sets.
Image type separation
To segregate the four image types (sticker, clinical, close-up, and dermoscopic), we utilized a convolutional neural network based on the EfficientNetB0 architecture, incorporating a human-in-the-loop system for quality assurance. The usage of active learning minimized labelling effort. This classifier achieved a balanced accuracy of 98.9% on a validation set of 500 images, after being trained on 1,000 manually labelled images. The result was subsequently verified by expert dermatologists.
Automatic patient identification
The sticker images could contain the patient information at any location of the picture. To automate the information extraction, we trained a YOLOv3 architecture23 to determine the size and location of the sticker in the image. We trained the architecture on a subset of 200 hand-labeled images and validated it on 50 images obtaining an mAP of 0.72. This architecture was then used to detect the stickers in the rest of the images (see Fig. 4).
Following the automated detection of sticker locations, we used Tesseract OCR24 to read the patient name and identifier. Not all images presented the same kind of stickers. In addition, there was handwritten information on some sticker images, so the results were revised by image readers after the process was finished. Finally, after all the sticker images had been correctly identified, we propagated the label information to the consecutive clinical, close-up, and dermoscopic pictures.
Linking images to patient reports
Patient names, identifiers, and image capture dates were used to assign one of the possible diagnostic categories, retrieved from several Excel files maintained by the doctors. Not all the images had a corresponding row in the diagnostic file since doctors would often take photos of lesions that were not clinically relevant and hence were of no interest for the construction of the dataset.
Finally, several readers revised each dermoscopic image to check the plausibility of the diagnostic category. Discrepancies were handled by multiple reader revisions and, if unanimous agreement amongst the reviewing panel was not achieved, they were discarded from the dataset. The metadata for each patient was gathered by the clinical practitioners at the time of capture.
Data Records
All data records of the BCN20000 dataset are released under a Creative Commons Attribution 4.0 International (CC-BY 4.0) license and are permanently accessible to the public through the Figshare repository with 10.6084/m9.figshare.2414002825. The dataset is partitioned into a training set comprising 12,413 images and a test set consisting of 6,533 images. Dermoscopic images and metadata are available for both sets. However, detailed diagnosis information is exclusively provided for the training set, encompassing eight distinct diagnostic categories.
Table 1 provides a comprehensive analysis of the training set, delineating the distribution of images stratified by diagnosis, sex, and anatomical location at both patient and lesion levels. The test set introduces an additional ‘unknown’ category, exclusive to it and absent from the training cohort. This category includes lesions that could not be definitively classified into the established categories, reflecting the complexity often encountered in clinical practice.
Significantly, the dataset includes images of lesions located in hard-to-diagnose areas, such as nails and mucosa. This aspect makes the BCN20000 dataset distinct from other more curated and publicly available datasets, as it more closely mirrors the challenges and diversity of actual clinical settings, which is lacking in current dermatoscopy datasets. The dermoscopy metadata encompasses patient age, biological sex, primary anatomic site of the lesion, date of capture, definitive lesion diagnosis, and designated data split.
Dataset format
The dataset is composed of 18946 dermoscopic images and a “.csv” file with comma-separated values containing the image name, label information, and metadata for all patients. Images are encoded in Joint Photography Expert Group (JPEG) format26, and they are 1024 by 1024 pixels each.
Dataset metadata
The dataset presents the following metadata:
-
BCN filename: Unique identifier for each dermoscopy image file.
-
Age approximation: Patient’s age quantized in 5-year intervals from 0 to 85.
-
Anatomical site of the legion: Categorizes the lesion’s body location into six areas: anterior torso, upper extremities, lower extremities, head/neck, palms/soles, and oral/genitalia.
-
Diagnosis: Specifies the lesion presented on the dermatoscopic image.
-
Lesion id: A unique identifier for each lesion, noting that some lesions may be represented by multiple images.
-
Capture date: The capture date of the photograph in YYYY-MM-DD format.
-
Sex: The patient’s sex, recorded as male, female, other, or not-reported.
-
Split: Designates the image’s allocation to either the test or training set.
Technical Validation
Histopathologic validation
Every melanoma diagnosis was confirmed through histopathologic examination of excised lesions by board-certified dermatopathologists, ensuring the highest level of diagnostic accuracy for melanoma classifications. Other malignant skin tumors also had histologic confirmation. Benign tumors were confirmed either histologically, through confocal examination, or by a clinical diagnosis from two experienced dermatologists at the referral center. Follow-up of tumors without excision confirmed stability to rule out malignancy.
Additional diagnostic modalities
Some lesions were diagnosed utilizing reflectance confocal microscopy, which offers near-cellular-level resolution27, or by digital dermoscopic follow-up to confirm stability. These methods provide a non-invasive, yet reliable means of identifying non-melanoma skin conditions.
Baseline CNN training
To assist researchers interested in automatic skin lesion classification of dermoscopy images, we developed and evaluated six baseline Convolutional Neural Networks (CNNs), which can serve as a starting point for future studies. The code can be found in the official repository at https://github.com/imatge-upc/BCN20000github.com/imatge-upc/BCN20000.
The implemented models are based on ResNet28 and EfficientNet29 architectures. We trained three small ResNet models with 18, 34, and 50 layers, respectively. From the EfficientNet architectures, we trained the B0, B1, and B2 models. We chose these architectures as they were used by the winners of the latest ISIC 2020 challenge18. All networks were pre-trained on ImageNet30. Our implementation uses the PyTorch framework for deep learning. We divided the data into training, validation, and testing with 75%, 5%, and 20% respectively.
During training, each RGB input image was resized to the appropriate model’s input size and augmented by performing the following random operations: resized crops with scales 0.8 to 1.2 and aspect ratio of 0.9 to 1.1, transforming from colour to grey-scale with 20% probability or applying permutation with 80% probability, including brightness (20%), saturation (20%), contrast (20%) and hue (20%), and horizontal and vertical flips (50%). Finally, the pixel values were normalized according to ImageNet mean and variance. In addition, we used weighted sampling to construct a uniform class distribution in the training batches to account for the severe class imbalance present in the dataset. We employed the Adam optimizer and used Cross-Entropy as the loss function throughout the training process. Finally, we trained the model for 130 epochs, or until the early stopping criterion was met, which involved halting training if the selected metric did not decrease for 20 consecutive epochs. The metric used to compare the models is balanced accuracy which measures the overall accuracy of a classification model by considering the proportion of true positive and true negative predictions for each class, making it desirable for unbalanced datasets. For the specific hyperparameters used during model training, refer to the official code repository https://github.com/imatge-upc/BCN20000.
Around 20% of the images in the dataset present a dark frame. This phenomenon would introduce biases in the models. We used a cropping algorithm to detect the Region Of Interest (ROI), and the lesion, and remove the dark background as much as possible. First, images were converted to grayscale and binarized with a low threshold, to retain the entire dermoscopy field of view. Then, we found the center of mass and the major and minor axis of an ellipse with the same second central moments as the ROI area. Next, based on these values, we derived a rectangular bounding box for cropping, covering the relevant field of view. Finally, we automatically determined the necessity of the cropping by comparing the mean intensity inside and outside the bounding box. Visual inspection showed that the method was robust. Examples of four different lesions before and after cropping are shown in Fig. 5. We fine-tuned the six pretrained architectures for the cropped and uncropped datasets and found a mean increase of 2.5% in balanced accuracy across all the architectures trained on the cropped dataset. Table 2 shows the balanced accuracy of each model for the cropped and uncropped datasets. The best results were obtained for the EfficientNet-B2 model on the cropped dataset, with a balanced accuracy of 0.461. The mean and variance were obtained using stratified k-fold cross-validation with five folds.
Usage Notes
The images in the BCN20000 dataset correspond to the following categories: nevus, melanoma, basal cell carcinoma, seborrheic keratosis, actinic keratosis, squamous cell carcinoma, dermatofibroma, vascular lesion and an extra OOD class only available on the test set. An example of each class on the training set can be found in Fig. 2. Each image is coupled with additional information regarding the anatomic location of the lesion, patient age and sex, image capture date, lesion ID, and diagnosis. The dataset was part of the ISIC 2019 and 2020 Challenge, where participants were asked to classify among various diagnostic categories and identify out of the distribution situations, where the algorithm is seeing a skin lesion it has not been trained to deal with.
Description of diagnosis categories
-
Melanoma (MEL): Melanoma is a malignant neoplasm derived from melanocytes that may appear in different variants. Melanomas can be invasive or non-invasive (in situ). We included all common variants of melanoma, such as melanoma in situ, superficial spreading melanoma, nodular melanoma, lentigo malignant melanoma, acral lentiginous melanoma and mucosal melanoma.
-
Nevus (NV): Melanocytic nevi are benign neoplasms of melanocytes and appear in many variants. The variants may differ significantly from a dermatoscopic point of view. However, in contrast to melanoma, they are usually symmetric with regard to the distribution of colour and structure in typical nevi and more irregular in atypical nevi.
-
Basal cell carcinoma (BCC): Basal cell carcinoma is a common variant of epithelial skin cancer that rarely metastasizes but grows destructively if untreated. It appears in different morphologic variants (flat, nodular, pigmented, cystic, infiltrative, morpheiform, etc).
-
Squamous carcinoma (SCC): It develops in the squamous cells that make up the middle and outer layers of the skin. Squamous cell carcinoma of the skin is usually not life-threatening, though it can be aggressive. Untreated, squamous cell carcinoma of the skin can grow large or spread to other body parts, causing severe complications.
-
Dermatofibroma (DF): Dermatofibroma is a benign skin lesion regarded as either a benign proliferation or an inflammatory reaction to minimal trauma. The most common dermatoscopic visual clue are reticular lines at the periphery with a central white patch denoting fibrosis.
-
Benign Keratosis (BKL): “Benign keratosis” is a generic class that includes seborrheic keratoses (“senile wart”), solar lentigo - which can be regarded as a flat variant of seborrheic keratosis - and lichen-planus like keratoses, which corresponds to a seborrheic keratosis or a solar lentigo with inflammation and regression. The three subgroups may look different dermatoscopically, but we grouped them because they are similar biologically and often reported histopathologically under the same generic term. From a dermatoscopic view, lichen planus-like keratoses are especially challenging because they can show morphologic features mimicking melanoma and are often biopsied or excised for diagnostic reasons. In addition, the dermatoscopic appearance of seborrheic keratoses varies according to anatomic site and type.
-
Actinic keratosis (AK): is a rough, scaly patch on the skin that develops from years of sun exposure. It’s often found on sun-exposed areas such as the face, lips, ears, forearms, scalp, neck, or back of the hands. Left untreated, the risk of actinic keratoses turning into a type of skin cancer called squamous cell carcinoma is about 5% to 10%.
-
Vascular (VASC): cutaneous vascular lesions are benign lesions which may mimic malignant skin tumours. Several vascular diseases are included in this class, such as capillary angioma, hemangioma, pyogenic granuloma, cavernous angioma, venous angioma, verrucous angioma, lobed angioma, dermal angioma, thrombosed angioma, venous lake, haemorrhoid, ectatic vascular structure, or thrombosed vascular structure.
Code availability
The code used for the pre-processing of the images and the training of the different architectures seen in Fig. 5 can be found at https://github.com/imatge-upc/BCN20000. All code is licensed under the Attribution 4.0 International (CC BY 4.0) License.
References
Tripp, M. K., Watson, M., Balk, S. J., Swetter, S. M. & Gershenwald, J. E. State of the science on prevention and screening to reduce melanoma incidence and mortality: The time is now. CA: a cancer journal for clinicians 66, 460–480 (2016).
Bleyer, A., Viny, A. & Barr, R. Cancer in 15-to 29-year-olds by primary site. The oncologist 11, 590–601 (2006).
Hanly, P., Soerjomataram, I. & Sharp, L. Measuring the societal burden of cancer: The cost of lost productivity due to premature cancer-related mortality in e urope. International journal of cancer 136, E136–E145 (2015).
Argenziano, G. et al. Dermoscopy of pigmented skin lesions: results of a consensus meeting via the internet. Journal of the American Academy of Dermatology 48, 679–693 (2003).
Kittler, H., Pehamberger, H., Wolff, K. & Binder, M. Diagnostic accuracy of dermoscopy. The lancet oncology 3, 159–165 (2002).
Johr, R. H. Dermoscopy: alternative melanocytic algorithms—the abcd rule of dermatoscopy, menzies scoring method, and 7-point checklist. Clinics in dermatology 20, 240–247 (2002).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115 (2017).
Xie, F. et al. Melanoma classification on dermoscopy images using a neural network ensemble model. IEEE transactions on medical imaging 36, 849–858 (2016).
Bi, L., Kim, J., Ahn, E. & Feng, D. Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. arXiv preprint arXiv:1703.04197 (2017).
Abbas, Q., Emre Celebi, M. & Fondón, I. Computer-aided pattern classification system for dermoscopy images. Skin Research and Technology 18, 278–289 (2012).
Barata, C., Celebi, M. E. & Marques, J. S. Improving dermoscopy image classification using color constancy. IEEE journal of biomedical and health informatics 19, 1146–1152 (2014).
Silva, C. S. & Marcal, A. R. Colour-based dermoscopy classification of cutaneous lesions: an alternative approach. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization 1, 211–224 (2013).
Hardie, R. C., Ali, R., De Silva, M. S. & Kebede, T. M. Skin lesion segmentation and classification for isic 2018 using traditional classifiers with hand-crafted features. arXiv preprint arXiv:1807.07001 (2018).
Marchetti, M. A. et al. Results of the 2016 international skin imaging collaboration isbi challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. Journal of the American Academy of Dermatology 78, 270 (2018).
Gutman, D. et al. Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (isbi) 2016, hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:1605.01397 (2016).
Codella, N. C. et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), 168–172 (IEEE, 2018).
Codella, N. et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:1902.03368 (2019).
Rotemberg, V. et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Scientific data 8, 34 (2021).
Tschandl, P. et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. The Lancet Oncology (2019).
Tschandl, P., Rosendahl, C. & Kittler, H. The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data 5, 180161 (2018).
Combalia, M. et al. Validation of artificial intelligence prediction models for skin cancer diagnosis using dermoscopy images: the 2019 international skin imaging collaboration grand challenge. The Lancet Digital Health 4, e330–e339 (2022).
ISIC. Isic challenge 2019. https://challenge2019.isic-archive.com/. [Online; accessed 2019-07-30] (2019).
Redmon, J. & Farhadi, A. Yolov3: An incremental improvement. arXiv preprint arXiv:1804.02767 (2018).
Google. Tesseract ocr (2019). https://opensource.google.com/projects/tesseract. [Online; accessed 2019-07-30].
Carlos Hernández-Pérez et al. Bcn20000: Dermoscopic lesions in the wild, Figshare https://doi.org/10.6084/m9.figshare.24140028.v1 (2023).
International Organization for Standardization. Digital compression and coding of continuous-tone still images - part 1: Requirements and guidelines. Standard ISO/IEC 10918-1:1992, ISO/IEC JTC 1 (1992).
Stevenson, A. D., Mickan, S., Mallett, S. & Ayya, M. Systematic review of diagnostic accuracy of reflectance confocal microscopy for melanoma diagnosis in patients with clinically equivocal skin lesions. Dermatology practical & conceptual 3, 19 (2013).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, 770–778 (2016).
Tan, M. & Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. International Conference on Machine Learning. PMLR, (2019).
Deng, J. et al. Imagenet: A large-scale hierarchical image database. In 2009 IEEE conference on computer vision and pattern recognition, 248–255 (Ieee, 2009).
Acknowledgements
We acknowledge the support of the International Skin Imaging Collaboration (ISIC). This research was supported by the Spanish Research Agency (AEI) under project PID2020-116907RB-I00 of the call MCIN/ AEI /10.13039/501100011033 and the project 718/C/2019 with id 201923-30 and 201923-31, funded by Fundació la Marató de TV3, iTOBOs grant from the European Union’s Horizon 2020 research and innovation programme num 965221. Other funding sources include the Melanoma Research Alliance Young Investigator Award 614197. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
Author information
Authors and Affiliations
Contributions
Conceptualization by J.M. and S.P.; methodology by J.M. and S.P.; software development by C.H. and M.C.; validation by J.M., V.V. and M.C.; formal analysis by J.M., C.H., S.Po., A.B. and V.V.; resources provided by A.H., J.M. and V.V.; writing–original draft preparation by C.H. and M.C.; writing–review and editing by C.H., M.C., A.B., S.Po., B.H., O.R., C.C., V.R., N.C., S.Po., S.P., A.H.J.M. and V.V.; funding acquisition by J.M., S.P., A.C.H. and V.V.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hernández-Pérez, C., Combalia, M., Podlipnik, S. et al. BCN20000: Dermoscopic Lesions in the Wild. Sci Data 11, 641 (2024). https://doi.org/10.1038/s41597-024-03387-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03387-w
- Springer Nature Limited