Abstract
Pluripotent stem cells (PSCs) provide unlimited resources for regenerative medicine because of their potential for self-renewal and differentiation into many different cell types. The pluripotency of these PSCs is dynamically regulated at multiple cellular organelle levels. To delineate the factors that coordinate this inter-organelle crosstalk, we profiled those long non-coding RNAs (lncRNAs) that may participate in the regulation of multiple cellular organelles in PSCs. We have developed a unique strand-specific RNA-seq dataset of lncRNAs that may interact with mitochondria (mtlncRNAs) and polyribosomes (prlncRNAs). Among the lncRNAs differentially expressed between induced pluripotent stem cells (iPSCs), fibroblasts, and positive control H9 human embryonic stem cells, we identified 11 prlncRNAs related to stem cell reprogramming and exit from pluripotency. In conjunction with the total RNA-seq data, this dataset provides a valuable resource to examine the role of lncRNAs in pluripotency, particularly for studies investigating the inter-organelle crosstalk network involved in germ cell development and human reproduction.
Similar content being viewed by others
Background & Summary
Long non-coding RNAs (lncRNAs) are transcripts of at least 200 nucleotides in length that lack a clear putative protein-coding ORF1. Although the number of characterized lncRNAs has dramatically increased, the biological roles of the lncRNAs in embryonic development, particularly in pluripotent reprogramming, have not been fully characterized. It was initially thought that lncRNAs were only present in the nucleus, but it is now clear that a number of lncRNAs encoded by the nucleus are also transported and localized to the cytoplasm2. LncRNAs found in the nucleus are usually related to epigenetic regulation at the transcriptional level. However, the finding that some lncRNAs are associated with cytoplasmic polyribosomes (prlncRNAs) suggests the coding potential3 for these RNAs. It is also possible that these cytoplasmic lncRNAs subserve a post-transcriptional regulatory function4 by fine-tuning the speed of translation or otherwise modifying the activity of ribosomes. Moreover, the base-pairing capability of prlncRNAs indicates that they can also interact with and regulate specific mRNAs5.
In addition to the numerous nuclear genome-encoded lncRNAs, the mitochondrial genome generates at least eight lncRNAs, several dsRNAs, and numerous small RNAs that either translocate into the cytosol and/or nucleus or remain in the mitochondria to perform various biological functions6. Three mitochondrial lncRNAs (mtlncRNAs), lncND5, lncCyt b, and lncND6, were identified using deep sequencing data from human HeLa cell mitochondria7. In both ischemic and non-ischemic human failing hearts, changes in the abundance of mtlncRNAs were noted in the left ventricle8. Mitochondrial function critically depends on the import of many nuclear-encoded macromolecules. In all eukaryotes, selected nuclear genome-encoded non-coding RNAs are partially redirected from the nucleus to the mitochondria, where they regulate mitochondrial gene expression9. These nuclear and mitochondrial genome-encoded lncRNAs may engage in inter-compartment crosstalk, either “nucleus-to-mitochondria” or “mitochondria-to-nucleus,” to maintain cellular homeostasis10,11. Aberrant shuttling of lncRNAs in this inter-compartment crosstalk is associated with human diseases, including cancer. For example, the mitochondria-encoded lncRNA lncCytB was located in mitochondria in normal hepatic cells. In hepatoma HepG2 cells, however, this lncRNA is considerably enriched in the nucleus12. In contrast, the nuclear genome-encoded lncRNA MALAT1 is aberrantly transported to the mitochondria, where it acts as an epigenetic regulator to control metabolic reprogramming in hepatoma cells13. Thus, some lncRNAs may act as vital epigenetic messengers to coordinate the inter-organelle crosstalk.
Mammalian embryonic stem cells (ESCs) originate from the ectoderm of developing embryos and can differentiate into three germ layers. Induced PSCs (iPSCs) are derived from the direct reconstitution of somatic cells into ESC-like pluripotent cells via the introduction of specific transcription factors. The use of ESC and iPSCs in clinical treatments for tissue repair has prompted in-depth research into their biological characteristics14. However, the molecular mechanisms underlying stem cell differentiation remain unknown, and research on lncRNAs may shed new light on this process15,16. Forty lncRNAs were identified in mouse ESC PSCs. After knocking out 30 of them, mESCs were induced to differentiate into distinct lineages17. Chakraborty et al. identified three lncRNAs that maintained the pluripotent stem cell characteristics in mESCs and dubbed them pluripotency-related non-coding transcripts 1–3 (Panct1–3). After knocking out Panct1, the expression of pluripotency markers was decreased while the expression of lineage markers was increased18. Loewer et al. discovered that iPSCs are abundant in the intergenic long-chain non-coding RNA (lincRNA) ST8SIA3, named lincRNA reprogramming regulator (linc-ROR), which promotes the formation of iPSC clones by inhibiting pro-apoptotic pathways19.
Our group previously published a combined pluripotency-associated lncRNA dataset that covers the data of RNA reverse transcription-associated trap sequencing (RAT-seq), chromatin RNA in situ reverse transcription sequencing (CRIST-seq), and RNA-seq20,21,22,23,24,25. The integration of these datasets allowed us to identify many differentially expressed lncRNAs that are not only associated with pluripotency but also function as chromatin factors to regulate pluripotency. These lncRNAs epigenetically coordinate the pluripotency-regulatory network and regulate stem cell fate through various epigenetic mechanisms, including coordinating intrachromosomal looping, recruiting methyltransferases and demethylases, and activating eRNA pathway of stemness genes20,21,22,23,24,25. Some lncRNAs, like nuclear Peln1, use a novel trans mechanism to regulate the exit from pluripotency21.
However, the role of the lncRNAs involved in the inter-organelle regulatory network, including nuclear-mitochondrial-polyribosomal crosstalk, has not been characterized. This data descriptor presents a unique strand-specific RNA-seq dataset of prlncRNA and mtlncRNA from human iPSCs, H9 ESCs, and fibroblasts. This dataset provides a valuable resource for studying these inter-organelle lncRNAs and should provide the means of examining mechanisms underlying the regulation of germ cell development and human reproduction. Most importantly, these mitochondrial and polyribosomal RNA-seq data and total RNA-seq data may help define those lncRNAs that determine stem cell fate by coordinating inter-organelle epigenetic regulatory networks.
Methods
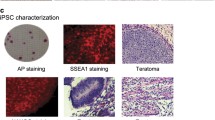
Characterization of iPSCs, H9 cells, and fibroblasts
Human embryonic stem cells (H9, WA09) were purchased from Wicell Research Institute (hPSCReg ID: WAe009-A). Skin fibroblasts (FBL, SPF7) were purchased from Coriell cell repository (AG06299) and cultured as described in previous studies26,27. Two iPSC cell lines (C11, S0730) were kindly provided by Professor Esteban of the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences. They were induced from human urinary fibroblast using lentiviruses carrying the OCT4, SOX2, KLF4, and c-MYC as previously described28. The pluripotency of the cultured human iPSC and H9 PSCs were examined by morphology (Fig. 1a) and positive immunostaining of stem cell markers OCT4, SOX2, and NANOG (Fig. 1b). The terminally differentiated status of human fibroblasts was confirmed by positive staining of vimentin (Fig. 1b). Specifically, cells were fixed with 4% paraformaldehyde/PBS for 10–15 min, rinsed with PBS, then permeabilized and blocked with 0.1% Triton X-100/PBS containing 3% BSA for 30 min. After washing with PBS, cells were incubated first with antibodies against OCT4 (Abcam, ab19857), SOX2 (Abcam, ab97959), and NANOG (Invitrogen, PA1-097) at 4 °C overnight, followed by Alexa Flour 555 labelled secondary antibody (Invitrogen, A-21429) staining. After washing three times with PBS, samples were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI, Invitrogen, D1306). Negative controls were stained without the use of primary antibodies. Fluorescence images were acquired with an Olympus FLUOVIEW FV3000. The pluripotency difference between the stem cells and fibroblasts was also confirmed by qPCR assay of stemness genes OCT4, SOX2, and NANOG (Fig. 1c).
Characterization of iPSCs, H9 cells, and fibroblasts. (a) Representative morphology of cell lines used for RNA-seq. (b) Characteristics of cells by immunofluorescent staining with antibodies against biomarkers. The pluripotency of iPSCs and H9 ESCs was characterized by the positive staining of OCT4, SOX2, and NANOG stem cell markers. The terminally differentiated status of fibroblasts (FBL) was identified by the positive staining of vimentin. (c) Quantitative analysis of pluripotent biomarkers OCT4, SOX2, and NANOG by RT-qPCR.
Sucrose gradient separation of polyribosomes and mitochondria
To isolate polyribosomes, cell lysates were prepared after 10 min of cycloheximide treatment at 37 °C to stabilize translating polysomes, and sucrose gradient separation and fractionation were performed as previously described (Fig. 2a)29. The polysome fractions determined by 260 nm absorbance were pooled for expression analysis (Fig. 2b).
Schematic diagram of sample collection and mitochondrial RNA purity. (a) The schematic diagram of sample collection and RNA sequencing. The polysomes were separated by sucrose density gradient centrifugation, the mitochondria were isolated using Qproteome Mitochondria Isolation Kit (Qiagen, USA), and total RNAs were extracted with Trizol reagent. The RNAs were then reverse-transcribed for library sequencing. (b) Polysome fractions were collected from each sample. A260 absorbance profiles were used to determine the polysome fractions in the sucrose density gradients. (c) Mitochondrial RNA purity was determined by the enrichment of nuclear-located RNA U2 and mitochondrial-located RNA COX2. The read counts in the RNA U2 and COX2 gene loci are shown by the bam files generated from the sequencing data.
Preparation of mitochondria
Mitochondria were prepared and purified using Qproteome Mitochondria Isolation Kit (Qiagen, USA) according to the manufacturer’s instructions. As previously reported30, 5 × 108 cells were suspended in lysis buffer, incubated in ice for 10 min, and centrifuged at 1000 × g for 10 min at 4 °C. The pellet was rewashed with lysis buffer and resuspended in disruption buffer, followed by passing 10 times through a 24-gauge needle to ensure complete cell disruption and centrifuged at 1000 × g. The supernatant was centrifuged at 8000 rpm for 10 min at 4 °C to pellet the mitochondria. The mitochondria were washed and purified by adding them on top of layers of purification and disruption buffers. The solution was centrifuged at 13000 rpm for 15 min at 4 °C. The mitochondrial ring at the interface of purification buffer/disruption buffer was collected and washed in mitochondria storage buffer.
The purity of the mitochondrial RNAs (mtRNAs) was reflected by the expression ratio of mitochondrial RNA COX2 and nuclear RNA U2. As shown in Fig. 2c, the read counts of COX2 in the three types of cells were significantly higher than those of the U2 RNA, and the expression ratio of COX2 and U2 was ~200–5000.
RNA extraction, cDNA library establishment, and Illumina sequencing
After pluripotency confirmation, Illumina RNA library sequencing was used to identify RNAs and lncRNAs that are differentially expressed in the reprogrammed cells (Fig. 2a). Total RNA was extracted using Trizol reagent (15596-018, Invitrogen, CA) according to the manufacturer’s instructions. The isolated RNAs were checked for RNA integrity by an Agilent Bioanalyzer 2100 (Agilent Technologies, CA, US). Total RNA was further purified by RNAClean XP Kit (A63987, Beckman Coulter, CA). RNase-Free DNase I (79254, QIAGEN, CA) was used to remove any contaminating DNA.
Ribosomal RNA was removed by a Ribo-Zero rRNA Removal Kit (#MRZH11124, Illumina, CA). RNAs were then fragmented into small pieces using a fragmentation reagent. The fragmented RNAs were subjected to first-strand cDNA synthesis using random hexamer-primed reverse transcription (18064014, SupperScript II reverse Transcriptase, Invitrogen, CA), followed by second-strand cDNA synthesis (Q32850, Qubit dsDNA HS Assay Kit, Invitrogen, CA). The cDNA fragments were 3′ adenylated and ligated with adaptors for PCR amplification for library construction. The library quality was assessed using Agilent2100. The libraries were clustered on an Illumina cBot Instrument and pair-sequenced using the Illumina NovaSeq 6000 platform.
Raw read filtering and transcript mapping
The raw sequencing reads were subjected to fastp v0.20.031 for removing: 1) adapter sequences in reads; 2) bases with a 3′ end Q value less than 20, indicating that the base error rate is greater than 0.01, where Q = −10logerror_ratio; 3) reads less than 25 in length; and 4) the ribosome RNA sequences of the species. The obtained clean reads were aligned to the human reference genome (GRCh38.p13) using the spliced mapping algorithm of StringTie32, which enables segmentation of reads that cannot be fully matched for mapping and is thus more suitable for eukaryotic transcriptome sequencing data containing intron regions. The alignments allowed for two mismatches; each read allowed for multiple hits < = 2, and the mapping generated BAM files.
The following software versions were used for quality control and data analysis: FastQC (v0.11.5): (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) QC filtering was performed using fastp 0.20.1 program with the default setting. (https://github.com/OpenGene/fastp). All reads were aligned to the human reference genome sequence by the STAR version 2.7.3a program. (https://github.com/alexdobin/STAR). Default parameters were used in the analyses.
LncRNA identification
To identify known lncRNA, we used the Ensembl gene and transcript sequences, which have comprehensive annotation and detailed classification information of lncRNAs. In addition, we integrated other databases to verify the reliability of the lncRNAs. To evaluate the coding potential, lncRNAs were filtered using four different coding potential prediction algorithms including “Coding Potential Calculator 2”, “Coding-Non-Coding Identifying Tool”, “Coding-Potential Assessment Tool”, and “CPPred”. The novel lncRNAs were identified by taking the intersection of these four algorithms.
Cuffcompare in cufflinks (version: 2.2.1)33 was used to compare the mapping-derived annotations to the reference annotations to obtain novel lncRNA transcripts that did not match known annotated genes. Three types of transcripts (I, u, and x) were further extracted for lncRNA prediction, where i indicates transfrags falling entirely within a reference intron, u indicates unknown and intergenic transcripts, and x refers to exonic overlap with reference on the opposite strand. Then, transcripts with a length greater than or equal to 200 bp, more than two exons, and ORF less than 300 were chosen. Pfam34, the Coding Potential Calculator (CPC)35, and the Coding-Non-Coding Index (CNCI)36 were used for prediction, and the intersection of their predicted results was obtained. After removing known lncRNA sequences, transcripts that were not significant compared to Pfam and had CPC and CNCI scores less than 0 were designated as potential novel lncRNAs. The numbers of known and novel lncRNA and mRNA transcripts detected by each dataset are shown in Table 1.
Expression abundance quantification
To standardize the expression level and make lncRNA expression levels between different samples comparable, we used cufflinks (version: 2.2.1)33 to convert the tophat mapping results to FPKM (Fragments Per Kilobase of Exon Model per Million Mapped Reads)37. The primary process is to obtain the precise location of the gene from the existing gene annotation file, then to count the reads covering the gene area, and finally to calculate the standardization of gene expression using the gene length and read count using the following formula:
where total exon fragments refer to the number of fragments aligned to the gene exon (fragment: a pair of reads), exon length refers to the total length of the gene exon, and mapped reads refer to the total number of reads aligned to the reference genome.
Differential expression analysis
Following quantification, the identification of differentially expressed lncRNAs (DE lncRNA) between different samples was performed using edgeR38, which can leverage the bootstraps of Kallisto to correct for technical variation. Multiple hypothesis testing was used to correct the obtained p-value, and the threshold was determined using False Discovery Rate39,40. The corrected p-value was then set as the q-value, and the statistical significance threshold was set to a q-value < = 0.05 (−log10 q-value > 1.3). Simultaneously, we calculated the differential expression fold change in terms of the FPKM value and set the biological significance threshold to a minimum of a two-fold change. As a result, we defined DE lncRNAs as those with biological and statistical significance. The data of DE analysis of mtlncRNAs versus RNA-seq and prlncRNAs versus RNA-seq are shown in Figure S1. The lncRNAs that are significantly enriched in mtlncRNAs and prlncRNAs compared with total RNAs have been deposited in GEO dataset GSE21668941.
Target gene prediction of DE lncRNAs
Since lncRNAs can regulate target gene expression at both the transcriptional and post-transcriptional levels, lncRNA target genes can be identified by analyzing the positional relationship (co-location) and expression correlation (co-expression) between lncRNAs and protein-coding genes42. The co-location method, for example, is based on the potential regulatory effect of lncRNA on nearby protein-coding genes. Therefore, target gene identification can be accomplished by searching for sequences within 100 kb upstream and downstream of lncRNA43. The co-expression analysis is based on the fact that certain lncRNAs can act on distant target genes. As a result, identifying its target genes is accomplished by correlating the expression of different gene products. Generally, this analysis is performed when the sample size exceeds five44. Due to the small sample size in this study, only the co-location prediction results are presented.
GO and KEGG enrichment analysis
Gene Ontology (GO) enrichment analyses of target genes of differentially expressed lncRNAs were implemented by the GOseq45,46. The specific principle is to map the selected DE lncRNA-targeted genes to each term of the GO database to calculate the number of genes contained in each entry. The hypergeometric test was then used to identify significantly enriched GO terms (with a corrected p-value < 0.05) enriched by DE lncRNAs-targeted genes. KEGG (http://www.genome.jp/kegg/) is a database resource for understanding high-level functions and utilities of a biological system, such as the cell, the organism, and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high throughput experimental technologies. We used KOBAS software47 to test the statistical enrichment of DE lncRNAs-targeted genes in KEGG pathways. The GO and KEGG enrichment of differentially-expressed RNA transcripts was deposited in NCBI GEO databases (GSE216689)41.
Data records
The sequencing data in the fastQ format have been deposited in NCBI GEO databases (GSE216689)41 The FastQ format data will serve as the raw sequencing data for further downstream processing. The processed data (bedgraph), the general transfer format (gtf) file, the FPKM values and the genome locations of all detected transcripts have been deposited in NCBI Gene Expression Omnibus (GSE216689)41.
Technical Validation
Quality control of RNA samples and library
The quality of RNA samples prepared from H9, iPSC, and fibroblasts was determined using the Agilent Bioanalyzer 2100 (Agilent Technologies). Each sample had an RNA integrity number greater than 7.0, indicating that the values met the requirements for an RNA-sequencing library. The library quality was checked using Agilent2100, producing an average of 370–380 bp fragments, including adapters.
To further validate the quality of these datasets, we compared the abundance of lncRNAs found in polyribosomes that have been reported in a colon cancer cell line4 and in a hepatocellular carcinoma cell line48. As seen from the FPKM data in Figure S2a,b, lncRNAs CASC7 and TUG1 were abundantly enriched in polyribosomal RNA-seq datasets as compared with the total RNA-seq dataset. Similarly, COX2 and ND5 were abundant in mitochondrial RNA-seq (Figs. S3a,b). In addition, we also used RT-qPCR to validate the abundance of these lncRNAs in isolated polyribosomal RNA and mitochondrial RNA samples (Figs. S4, S5).
Quality control of sequencing data and DE lncRNAs
We applied FastQC v0.11.5 software to determine sequencing data quality. The per base sequence quality was high, with a median quality score above 30. The pattern of GC composition was similar to the theoretical distribution, indicating that the samples were free from contamination. In addition, the sequence length distribution also corresponded to the theoretical curve. The sequencing on Illumina NovaSeq 6000 generated mitochondria-associated RNA raw reads and polyribosome-associated RNA raw reads for H9, iPSC, and fibroblasts, respectively. After removing low-quality reads, clean reads were obtained for H9 (87,491,656), iPSC (83,502,776), and fibroblast (109,026,364) mitochondria-associated RNAs, respectively. At the same time, polyribosome-associated RNA clean reads were also obtained for H9 (72,565,674), iPSC (67,917,382), and fibroblasts (89,788,274). After Seqtk filtering, a total of 84,225,062 (96.27%), 79,386,203 (95.07%) and 99,030,086 (90.83%) clean reads were generated for H9, iPSC, and fibroblast mitochondria-associated RNA, as well as 9,479,552 (13.06%), 40,462,642 (59.58%), and 79,180,606 (88.19%) clean reads for H9, iPSC, and fibroblasts polyribosome-associated RNA (Fig. 3a). These reads were then mapped to the human genome (GRCh38.p13) for lncRNAs using the STAR software49.
Differential expression analysis of RNA sequencing. (a) Quality control of RNA-sequencing data. (b) Principal component analysis (PCA) of transcripts in RNA-sequencing. (c) Heat map of transcript level correlation distance between samples. (d) Expression levels of protein-coding, known lncRNA, and novel lncRNA transcripts.
To evaluate between-group differences and within-group sample duplication, we conducted principal component analysis (PCA) (Fig. 3b). PCA is a mathematical dimensionality reduction process that uses an orthogonal transformation to convert a set of linearly related variables into a set of linearly uncorrelated new variables, also known as principal components, to display the data in a lower dimension feature. It is possible to maintain as much information as possible in the variables and limit the number of variables as little as possible by using PCA, simplifying both the calculation and the interpretation of the findings. Additionally, the PCA analysis can be utilized to identify the main component with the greatest contribution as the data representative for the results visualization.
Based on quantitative and differential expression analyses, Pearson’s correlation coefficients of the transcript expression level of each sample showed that H9 and C11 iPS cells had high similarity in transcript expression, while FBL cells had significant differences from the two kinds of pluripotent cells (Fig. 3c). The expression levels of different kinds of transcripts, including protein-coding, known lncRNA, and novel lncRNA, are shown in Fig. 3d.
Identification of novel pluripotency-associated polysomal lncRNAs
By integrating the polyribosomal RNA-seq and total RNA-seq data, we identified 11 novel lncRNAs from the top differentially expressed transcripts that were upregulated in both pluripotent stem cells H9 and C11. These RNA transcripts did not have known gene IDs and gene names, and they had higher FPKM in H9 and iPSC prRNAs than that in FBL. They were thus named PARIT (pluripotency-associated ribosome-interacting transcripts) 1–11 (Table 2). These lncRNAs were among the most upregulated prRNA transcripts between iPSC and FBL, as well as between H9 and FBL.
We then used RT-qPCR to confirm differential expression of these lncRNAs in the polyribosome fraction between H9/C11 and FBL cells (Fig. 4a) using specific primers (Table 3). The correlation between PARIT1-11 expression and stem cell pluripotency was validated by a cell differentiation test in C11 cells, introduced by replacing 20% of the supplement in the mTESR1 medium with FBS. The expression of PARIT1-11 was reduced with the addition of FBS, as were the stemness genes of OCT4, SOX2 and NANOG, as shown in Fig. 4b,c. Currently, we know very little about the function of these prlncRNAs and mtlncRNAs. Future studies are needed to explore the role of these lncRNAs using organelle-specific targeting approaches.
Identified novel lncRNAs that are associated with pluripotency. (a) RT-qPCR quantitation of lncRNAs PARIT1-11 in the polyribosome fraction between iPSCs and H9 cells and fibroblasts. Data are standardized over internal control β-Actin and are calculated as fold-changes of iPSCs and H9 cells over FBL. The experiment was performed in duplicate. (b) Downregulation of PARIT1-11 in cell differentiation. iPSCs and H9 cells were cultured with mTeSR1 supplemented (100%) according to the manufacturer’s instructions to maintain stemness or cultured with 80% supplement and 20% FBS to induce cell differentiation. The experiment was performed in duplicate. *p < 0.05 and **p < 0.01 compared with the 100% supplement group. (c) Downregulation of stemness genes in cell differentiation. The stemness genes OCT4, SOX2 and NANOG were detected by RT-qPCR during cell differentiation induced by culturing with 80% supplement and 20% FBS. The experiment was performed in duplicate. *p < 0.05 and **p < 0.01 compared with 100% supplement group.
Code availability
No custom code was generated for this work.
References
Mattick, J. S. & Rinn, J. L. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol 22, 5–7, https://doi.org/10.1038/nsmb.2942 (2015).
Carlevaro-Fita, J., Rahim, A., Guigo, R., Vardy, L. A. & Johnson, R. Widespread localisation of lncRNA to ribosomes: Distinguishing features and evidence for regulatory roles. bioRxiv, 013508 https://doi.org/10.1101/013508 (2015).
Ruiz-Orera, J., Messeguer, X., Subirana, J. A. & Alba, M. M. Long non-coding RNAs as a source of new peptides. Elife 3, e03523, https://doi.org/10.7554/eLife.03523 (2014).
van Heesch, S. et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol 15, R6, https://doi.org/10.1186/gb-2014-15-1-r6 (2014).
Pircher, A., Gebetsberger, J. & Polacek, N. Ribosome-associated ncRNAs: an emerging class of translation regulators. RNA Biol 11, 1335–1339, https://doi.org/10.1080/15476286.2014.996459 (2014).
Liu, X. & Shan, G. Mitochondria Encoded Non-coding RNAs in Cell Physiology. Front Cell Dev Biol 9, 713729, https://doi.org/10.3389/fcell.2021.713729 (2021).
Rackham, O. et al. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17, 2085–2093, https://doi.org/10.1261/rna.029405.111 (2011).
Yang, K. C. et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 129, 1009–1021, https://doi.org/10.1161/CIRCULATIONAHA.113.003863 (2014).
Jeandard, D. et al. Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches. Cells 8 https://doi.org/10.3390/cells8030286 (2019).
Dong, Y., Yoshitomi, T., Hu, J. F. & Cui, J. Long noncoding RNAs coordinate functions between mitochondria and the nucleus. Epigenetics & chromatin 10, 41, https://doi.org/10.1186/s13072-017-0149-x (2017).
Zhao, Y., Sun, L., Wang, R. R., Hu, J. F. & Cui, J. The effects of mitochondria-associated long noncoding RNAs in cancer mitochondria: New players in an old arena. Critical reviews in oncology/hematology 131, 76–82, https://doi.org/10.1016/j.critrevonc.2018.08.005 (2018).
Zhao, Y. et al. Aberrant shuttling of long noncoding RNAs during the mitochondria-nuclear crosstalk in hepatocellular carcinoma cells. Am J Cancer Res 9, 999–1008 (2019).
Zhao, Y. et al. Nuclear-encoded lncRNA MALAT1 epigenetically controls metabolic reprogramming in hepatocellular carcinoma cells through the mitophagy pathway. Mol Ther Nucleic Acids 23, 264–276, https://doi.org/10.1016/j.omtn.2020.09.040 (2021).
Nestor, M. W. & Noggle, S. A. Standardization of human stem cell pluripotency using bioinformatics. Stem Cell Res Ther 4, 37, https://doi.org/10.1186/scrt185 (2013).
Ghosal, S., Das, S. & Chakrabarti, J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev 22, 2240–2253, https://doi.org/10.1089/scd.2013.0014 (2013).
Huo, J. S. & Zambidis, E. T. Pivots of pluripotency: the roles of non-coding RNA in regulating embryonic and induced pluripotent stem cells. Biochim Biophys Acta 1830, 2385–2394, https://doi.org/10.1016/j.bbagen.2012.10.014 (2013).
Guttman, M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300, https://doi.org/10.1038/nature10398 (2011).
Chakraborty, D. et al. Combined RNAi and localization for functionally dissecting long noncoding RNAs. Nat Methods 9, 360–362, https://doi.org/10.1038/nmeth.1894 (2012).
Loewer, S. et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42, 1113–1117, https://doi.org/10.1038/ng.710 (2010).
Du, Z. et al. Combined RNA-seq and RAT-seq mapping of long noncoding RNAs in pluripotent reprogramming. Sci Data 5, 180255, https://doi.org/10.1038/sdata.2018.255 (2018).
Wang, Y. et al. Pluripotency exit is guided by the Peln1-mediated disruption of intrachromosomal architecture. J Cell Biol 221 https://doi.org/10.1083/jcb.202009134 (2022).
Du, Z. et al. Chromatin lncRNA Platr10 controls stem cell pluripotency by coordinating an intrachromosomal regulatory network. Genome Biol 22, 233, https://doi.org/10.1186/s13059-021-02444-6 (2021).
Jia, L. et al. Oplr16 serves as a novel chromatin factor to control stem cell fate by modulating pluripotency-specific chromosomal looping and TET2-mediated DNA demethylation. Nucleic Acids Res 48, 3935–3948, https://doi.org/10.1093/nar/gkaa097 (2020).
Wang, C. et al. Genome-wide interaction target profiling reveals a novel Peblr20-eRNA activation pathway to control stem cell pluripotency. Theranostics 10, 353–370, https://doi.org/10.7150/thno.39093 (2020).
Zhang, S. et al. Profiling the long noncoding RNA interaction network in the regulatory elements of target genes by chromatin in situ reverse transcription sequencing. Genome Res 29, 1521–1532, https://doi.org/10.1101/gr.244996.118 (2019).
Zhang, H. et al. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell 13, 30–35 S1934-5909(13)00205-1 [pii] https://doi.org/10.1016/j.stem.2013.05.012 (2013).
Chen, X. et al. Valproic Acid Enhances iPSC Induction From Human Bone Marrow-Derived Cells Through the Suppression of Reprogramming-Induced Senescence. J Cell Physiol 231, 1719–1727, https://doi.org/10.1002/jcp.25270 (2016).
Zhou, T. et al. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol 22, 1221–1228, https://doi.org/10.1681/ASN.2011010106 (2011).
Masek, T., Valasek, L. & Pospisek, M. Polysome analysis and RNA purification from sucrose gradients. Methods Mol Biol 703, 293–309, https://doi.org/10.1007/978-1-59745-248-9_20 (2011).
Sripada, L. et al. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One 7, e44873, https://doi.org/10.1371/journal.pone.0044873 (2012).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890, https://doi.org/10.1093/bioinformatics/bty560 (2018).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295, https://doi.org/10.1038/nbt.3122 (2015).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578, https://doi.org/10.1038/nprot.2012.016 (2012).
Sun, L. et al. Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinformatics 13, 331, https://doi.org/10.1186/1471-2105-13-331 (2012).
Kong, L. et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35, W345–349, https://doi.org/10.1093/nar/gkm391 (2007).
Sun, L. et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41, e166, https://doi.org/10.1093/nar/gkt646 (2013).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628, https://doi.org/10.1038/nmeth.1226 (2008).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, https://doi.org/10.1093/bioinformatics/btp616 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300, https://doi.org/10.1111/j.2517-6161.1995.tb02031.x (1995).
Benjamini, Y. & Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics 29, 1165–1188, 1124 (2001).
Hu, J., Zhou, L., Esteban, M. A. & Cui, J. NCBI Gene Expression Omnibus GSE216689 https://identifiers.org/geo/GSE216689 (2023).
Schmitt, A. M. & Chang, H. Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 29, 452–463, https://doi.org/10.1016/j.ccell.2016.03.010 (2016).
Bao, Z. et al. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res 47, D1034–D1037, https://doi.org/10.1093/nar/gky905 (2019).
Kopp, F. & Mendell, J. T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407, https://doi.org/10.1016/j.cell.2018.01.011 (2018).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11, R14, https://doi.org/10.1186/gb-2010-11-2-r14 (2010).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361, https://doi.org/10.1093/nar/gkw1092 (2017).
Bu, D. et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res 49, W317–W325, https://doi.org/10.1093/nar/gkab447 (2021).
Zhao, Y. et al. Nuclear-Encoded lncRNA MALAT1 Epigenetically Controls Metabolic Reprogramming in HCC Cells through the Mitophagy Pathway. Mol Ther Nucleic Acids 23, 264–276, https://doi.org/10.1016/j.omtn.2020.09.040 (2021).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, https://doi.org/10.1093/bioinformatics/bts635 (2013).
Acknowledgements
This work was supported by the National Key R&D Program of China (2020YFA0707704 and 2018YFA0106902), the Innovative Program of National Natural Science Foundation of China (82050003), the National Natural Science Foundation of China (82371872, 32000431, 81874052, 82301885), Fund of Jilin Provincial Science and Technology Department (YDZJ202301ZYTS003, 20200602032ZP, YDZJ202202CXJD004, and 20210303002SF), Youth Fund of the First Hospital of Jilin University (JDYY14202303), Fund of Jilin Province Labor Resources and Social Security Department (2023RY03), Fund of Jilin Provincial Development and Reform Commission (2021C010), and Fund of Changchun Science and Technology Bureau (21ZGY28).
Author information
Authors and Affiliations
Contributions
Lei Zhou: Investigation, project administration, conceptualization, manuscript draft writing, funding acquisition, validation, and resources. Hui Li: Experimental assays, cell culture, formal Analysis, and data curation. Tingge Sun: Investigation, methodology, formal analysis, data curation. Xue Wen: Investigation, methodology, formal Analysis, data curation. Chao Niu: Data curation and validation. Min Li: Data curation and validation. Wei Li: Resources, supervision, and funding acquisition. Miguel A. Esteban: Resources, supervision, and funding acquisition. Andrew R. Hoffman: Project supervision, manuscript review, and editing. Ji-Fan Hu: Project supervision, investigation, funding acquisition, project administration, and manuscript writing and editing. Jiuwei Cui: Conceptualization, supervision, funding acquisition, project administration and supervision, and manuscript review.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, L., Li, H., Sun, T. et al. Profiling mitochondria-polyribosome lncRNAs associated with pluripotency. Sci Data 10, 755 (2023). https://doi.org/10.1038/s41597-023-02649-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02649-3
- Springer Nature Limited