Abstract
Surface electromyogram (sEMG) offers a rich set of motor information for decoding limb motion intention that serves as a control input to Intelligent human-machine synergy systems (IHMSS). Despite growing interest in IHMSS, the current publicly available datasets are limited and can hardly meet the growing demands of researchers. This study presents a novel lower limb motion dataset (designated as SIAT-LLMD), comprising sEMG, kinematic, and kinetic data with corresponding labels acquired from 40 healthy humans during 16 movements. The kinematic and kinetic data were collected using a motion capture system and six-dimensional force platforms and processed using OpenSim software. The sEMG data were recorded using nine wireless sensors placed on the subjects’ thigh and calf muscles on the left limb. Besides, SIAT-LLMD provides labels to classify the different movements and different gait phases. Analysis of the dataset verified the synchronization and reproducibility, and codes for effective data processing are provided. The proposed dataset can serve as a new resource for exploring novel algorithms and models for characterizing lower limb movements.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Background & Summary
Intelligent human-machine synergy systems (such as rehabilitation equipment1, active prostheses2,3,4,5, and exoskeletons6) have developed rapidly with the continuous advancement in the medical, robotics, and computing fields. A key driver of these systems is the surface electromyogram (sEMG) signals acquired non-invasively that can be observed prior to the initiation of certain muscle-driven movements7. Thus, sEMG has been widely used in a broad spectrum of biomedical and non-biomedical applications that require the prediction of human motion intention8. For upper limb movement intent characterization, the Ninapro repository9,10 (a publicly accessible database) provides a considerable amount of datasets, including sEMG, joint angle (kinematics), and force (kinetics) data with corresponding labels to support the research community. Similarly, there are a few publicly available sEMG datasets for lower limb movement characterization, which have helped advance research and development to an extent in the field of rehabilitation robotics and the likes11,12,13,14,15,16,17. However, these lower limb sEMG datasets are limited in a number of ways that preclude their wide usage by researchers in the field. For instance, the currently available lower limb datasets do not directly provide detailed gait phase labels, which are essential for conducting appropriate investigations on gait phase characterization. Although gait phases can be computed from a few of the available datasets15,16,17 that include heel strike and toe-off information, this typically requires complex processing steps. At the same it is difficult to ensure fairness in the comparison of the accuracy of some machine learning algorithms due to the non-directly available labels for the gait phases. In addition, some of the existing lower limb datasets only considered recording data while subjects walk at various speeds on stairs or ground terrains11,12,13, without involving discrete movements that are needed in rehabilitation research and development18,19. Based on these reasons and the limitation of the existing publicly available lower limb datasets highlighted in Table 1, there is a need to provide a more comprehensive public lower limb datasets that could advance research, development, and innovation in the related domains. Table 1 presents the research demand versus characteristics of the available datasets, highlighting the drawbacks of the existing datasets, which the current study seeks to address.
To address the limitations of the available datasets, we designed an experiment that allowed the simultaneous acquisition of nine channels of sEMG, joint angle (kinematics), joint torque (kinetics) along with their corresponding labels from 40 healthy subjects who performed 16 different lower limb movements, and the obtained dataset is named as Shenzhen Institute of Advanced Technology Lower Limb Motion Dataset (SIAT-LLMD)20. In addition, codes that allow reading the data, pre-processing of sEMG, splitting of sEMG into analysis windows of various sizes, extraction of feature sets, normalization of extracted features, generation of sample data, and making log files are provided for complete handling of the data. In summary, this work provides a dataset (SIAT-LLMD)20 with additional types of movements and unified labels to promote the advancement of scientific research and comparison of the related algorithms in the field of lower limb movement characterization.
Methods
Participants
In this study, a total of 40 healthy adult subjects including 30 males and 10 females were recruited for the collection of the sEMG, kinematics, and kinetics data associated with multiple classes of lower limb movements. In order to protect the identity information of the subjects, we coded their names as Sub01 – Sub40. The average age across subjects is 24.5 years old (with a minimum and maximum age of 19 and 33, respectively); the average weight of the subjects is 63.8 Kg (with a minimum and maximum weight of 46.3 Kg and 85 Kg, respectively); the average height of the subjects is 1693 mm (with a minimum and maximum height of 1550 mm and 1820 mm, respectively); the average thigh length of the subjects is 391.4 mm (with a minimum and maximum thigh length of 325 mm and 455 mm, respectively); the average calf length of the subjects is 409.1 mm (with a minimum and maximum calf length of 355 mm and 477.5 mm, respectively); and the average foot length of the subjects is 224.2 mm (from the heel to the toe’s first phalangeal joint, with a minimum and maximum foot length of 200 mm and 255 mm, respectively). Meanwhile, a detailed description of each subject’s characteristics is presented in Table 2.
The participants were recruited from the Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences (SIAT-CAS) and the Shenzhen University via a publicity made on a social media application (specifically, the WeChat platform), describing the goal, experimental setup, and requirements of the study. Interested participants that signed up to take part in the study were given further descriptions (using photos and videos, etc.) about the goal and experimental procedures to ensure that they fully understood before participating in the study. Also, the participants were given the chance to ask related questions and responses were administered accordingly to ensure adequate understanding before the data collection began. The experimental design and protocol were reviewed and approved by the ethics committee led by the Institutional Review Board (IRB) of SIAT-CAS with an approval number of SIAT-IRB-210315-H0555.
Experimental setup and Equipment
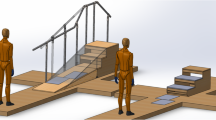
A motion capture system with six cameras (Eagle, Motion Analysis, USA) was used to collect the original kinematic data; two six-dimensional force platforms (OR6-7, AMTI, USA) were used to record the ground reaction force (GRF); and a wireless sEMG acquisition system (DELSYS, USA) employed for the recording of the sEMG data (Fig. 1).
The motion capture system was set at a sampling frequency of 60 Hz to track the three-dimensional (3D) trajectories of a set of configured 41 cutaneous reflective markers as shown in Fig. 2. This set of markers was used in the ‘Gait 2392’ OpenSim (a software from the National Center for Simulation in Rehabilitation Research) model21. The design of the markers is referred to in the guide for the usage of the Motion Capture and Analysis System and the recommendations of the International Society of Biomechanics22,23. The key markers in the bones used for the inverse kinematics calculation were placed based on guide from two skilled experimenters using the anatomical palpation method. The other markers used as redundancy for calculating missed markers were placed by the experimenters based on their personal experience. These redundant markers have zero weight during the calculation process. At the same time, to ensure the accuracy of the markers’ position, the subjects were asked to wear a light T-shirt, tight-fitting experimental shorts, take off their shoes, wear an experimental cap, and tuck their shirt into the shorts. Then, the T-shirts were wrapped with self-adhesive tensioners, and the subjects were instructed not to adjust the clothing midway into the experiment to ensure conformity with the protocol.
The ground reaction force (GRF) was recorded by the force platforms of 508 mm length and 464 mm breadth sampled at 1920 Hz, placed at the same height as the ground except when the subjects went upstairs or downstairs. For the stairs scenes, a stairs bracket that enables the force platforms to be set up in a desired manner (as shown in Fig. 1b) was designed. After setting up the platform, the height, width, and length of the stair are 150 mm, 260 mm, and 470 mm, respectively.
Thereafter, a total of nine-channel sEMG signals were collected by using a wireless acquisition system sampled at 1926 Hz, and the electrodes were firmly fixed on the tensor fascia lata, rectus femoris, vastus medialis, semimembranosus, upper tibialis anterior, lower tibialis anterior, lateral gastrocnemius, medial gastrocnemius, and soleus muscles on the left leg with a double-sided tape. During the experiment, we ensured that suggestions provided by the Institute of Neurology, Department of Clinical Neurophysiology were adhered to24. Before the placement of electrodes, the skin surfaces of the subjects were cleaned with alcohol cotton pads containing 75% alcohol to remove skin oil to avoid electrode impedance issues that may affect the quality of the recorded signals during the experiment. In addition, to avoid highly noisy signals that may result from vibration during the experiments, self-adhesive medical bandages were used to wrap and reinforce the EMG electrodes.
The whole system can collect the 3D trajectories of the marker set, GRF, and sEMG data, based on which, the kinematic and kinetic data can be calculated. Experimental data were collected after wearing the equipment (Fig. 1c). To ensure that the data is recorded appropriately, the reflective markers and sEMG sensors were carefully checked after each movement was performed by the subjects.
Experimental protocol
A total of 16 different lower limb movement tasks (including 12 different movements, no movement also known as the static state, and 3 different gaits) were considered in the data acquisition process. Precisely, these tasks include static (standing upright without making any movement), walking on a level ground, standing up, sitting down, walking upstairs, walking downstairs, knee lift, tipping the toe, leg lift forward, leg lift backward, knee lift then calf lift forward, leg lift sideward, heel strike, toe-off, lunging forward, and lunging backward (See the attached video and Table 3 for a clearer understanding of the experimental procedure for the tasks performed). According to the discrete nature of the movements, two methods (denoted as, method-1 and method-2) were employed for the data collection process which are represented in a flowchart shown in Fig. 3. In method-1, each participant is asked to perform a specific movement, and then repeat that same task nine times for gait phase related tasks and four times for the other tasks. In other words, each repetition constitute a raw data file, thus yielding a total of ten or five different raw data files per movement and subject. In contrast, for method-2, each participant is expected to perform a specific movement task five consecutive times in a row, yielding a single raw data file for the five trials. It is worth noting that method-1 was employed in collecting the data for static, walking on level ground, standing up, sitting down, stairs ascent, and stairs descent tasks because these tasks are designed to begin and terminate at certain intervals considering the experimental environment. Also, in method-2, the participants were required to follow an electronic metronome (1 second a beat, 8 seconds a loop) to complete each group of movements in 5 seconds and followed by 3 seconds of rest.
The summary for the justification on why we selected the movements considered in our proposed dataset is given as follows:
Goal one: The movements in SIAT-LLMD should include the main functions of the lower limb motion during daily life activities such as WAK, UPS, DNS, STDUP, and SITDN. In addition, in cases where an individual needs to make a sidestep in a certain direction, then LLF, LLB, and LLS movements will be needed.
Goal two: The SIAT-LLMD should be able to offer support for research involving a single gait phase in a gait cycle. For instance, when an individual starts walking from standing, the movement of the front leg corresponds to the HS, and the back leg corresponds to the TO. The KLCL is designed to support taking longer strides. At the same time, the longer strides require lunge to heel strike while crossing a ditch, so we considered the LUGF and LUGB movements in our dataset. In addition, the calf is often lifted when climbing up stairs which led us to include KLFT movement.
Goal three: It is also important to include movements associated with the angle range of motion of the three joints (hip, knee, and ankle) as much as possible to comprehensively support the research of using sEMG to track joint motion. To achieve this goal, we chose the KLFT, LLB, and LLS to cover the angle range of the hip, KLFT to cover the angle range of the knee, TPTO for the angle joint, and lunge movement (LUGF and LUGB) for the angle with stretch.
The above-described experimental protocol was carried out after the equipment was successfully set up. And the entire experiment lasted for 90 to 140 minutes per subject, depending on the learning rate and required rest time of each subject while about 40 minutes was spent wearing the markers and delsys sensors. In addition, subjects were permitted to stop the experiment and take adequate rest whenever they felt fatigued.
Data processing
The required Motion capture files can be obtained from the process mentioned in the previous section. Afterward, the following data processing procedures were performed: markers data processing, data extraction, kinematics and kinetics data calculation, and data alignment.
-
(1)
Markers data processing: The missing markers were fixed and named in accordance with the guidance presented in Fig. 2. These processes were carried out using the Cortex Software (Motion Analysis Company, USA). The function of “Rigid Body Join”, which ensures the correctness of the data by using the correct markers to fix the missing markers belonging to the same rigid, was preferred while fixing the missing markers. In addition, the smooth function was used on some frames following the suggestions from the Cortex Software, and a ‘.c3d’ file was finally generated as the output.
-
(2)
Data extraction: From the obtained ‘.c3d’ file, the sEMG data, ‘.trc’ file, and ‘.mot’ files were individually extracted. Meanwhile, the ‘.trc’ file and ‘.mot’ files were then used in the OpenSim software for calculating the corresponding kinematics and kinetics information.
-
(3)
Kinematics and Kinetics computation: Firstly, the STC and body weight data were used to scale the standard model for each subject. Secondly, the scaled model and the ‘.trc’ file were used to calculate the kinematics data associated with the joints. Thirdly, the ‘.mot’ file and kinematics data were used to calculate the kinetics associated with the joints, and the GRF data while the filter was set to 15 Hz.
-
(4)
Data alignment: Firstly, the kinematics and kinetics datasets were smoothed by the ‘smoothdata’ function (method: ‘sgolay’ window: 4) in Matlab. Secondly, the EMG data was used as a reference to upsample the kinematic and kinetic datasets. Finally, the sEMG, kinematics, and kinetic datasets were combined into a ‘.csv’ file and designated as the final data file.
-
(5)
Data trim and merge: For each of the STDUP and SITDN, the five data files were sequentially concatenated into one data file to keep the storage format consistent. Data lacking full GRF information were trimmed for the movements UPS, and DNS. The ten data files were concatenated into one data file for each WAK, UPS, and DNS, with the lift-foot-first group in the front and the right-foot-first group in the back.
Although sEMG signals usually require filtering before use, some studies still expect the direct application of raw data. So, raw EMG data and a set of codes have been provided in this work to enable easy exploration of our dataset by interested researchers. The codes provide different filters, normalization methods, and window settings that are suitable for the dataset, by virtue of which, researchers can easily obtain data that could be used for various analyses when considering statistical, machine learning, and other methods. It should be noted that the data lacking full GRF information in WAK is marked as NAN (Not a Number), and the NAN data needs to be excluded before using the enclosed codes to automatically process it. Besides, interested researchers may also use the data without excluding the NAN aspect but would need to either modify our codes or use a custom built codes.
Label
This data set provides two types of labels. The first type is suitable for STC, STDUP, SITDN, KLFT, TPTO, LLF, LLB, LLS, KLCL, HS, TO, LUGF, LUGB, while the second type is suitable for WAK, UPS, and DNS. In addition, the code in this work could be used to extract samples with the required labels or one-hot labels for researchers expecting to do pattern recognition for specific movement classification.
The first type of label includes ‘A’ and ‘R’ (Fig. 4), corresponding to the active or rest status of the subject with respect to their lower limb movements. These labels are determined by the force platforms (described in ‘Experimental setting and Equipment’ section) and kinematics data.
The second type of label marks the different gait phases (Fig. 4). Thus, there are five key gait events in the level walking scenario which include heel strike (HS), maximum stance flexion (MSF), maximum stance extension (MSE), toe-off (TO), and maximum swing flexion (MWF)25,26,27, three of which include HS, TO, and MWF, can be detected when going upstairs and downstairs, which were considered in our study. However, since the time from TO to MWF is short during stairs descent (DNS), the entire swing period is counted as the same state. MSF and MSE represent the moments during the stance phase when the knee flexion and extension angles respectively reach their maximum values. MWF refers to the moment of maximum knee flexion during the swing phase. The force platforms and kinematics data were used to detect these events, and then the number of labels were used to distinguish the data between the two consecutive gait events.
Data Records
The SIAT-LLMD includes the kinematic, kinetic, sEMG dataset, the details of the subjects, photos of experiments, and corresponding labels, which were recorded as shown in Fig. 5. The subjects’ basic information is contained in a file denoted as ‘SubjectsInformation.xlsx’ which holds the subject ID, age, body weight, and body size of each subject. Besides, each subject’s data is organized in a single folder that contains 16 data files with a naming format of ‘Subxx_xxx_data.csv’. In each data file, the first column holds time; kinematic data are recorded in the second to ninth column; the kinetic data are recorded in the tenth to seventeenth column; and the sEMG data are recorded in the eighteenth to twenty-sixth column. In addition, there is a clear header marked with the specific name of each kind of data in the file for easy understanding. The kinematic data from left to right are the joint angle of left hip adduction, left hip flexion, left knee flexion, left ankle flexion, right hip adduction, right hip flexion, right knee flexion, and right ankle flexion, respectively. The kinetic data from left to right are the joint torque of left hip adduction, left hip flexion, left knee flexion, left ankle flexion, right hip adduction, right hip flexion, right knee flexion, and right ankle flexion, respectively. And the sEMG data from left to right are the data collected from the tensor fascia lata, rectus femoris, vastus medialis, semimembranosus, upper tibialis anterior, lower tibialis anterior, lateral gastrocnemius, medial gastrocnemius, and soleus muscles in the left leg, respectively. The corresponding labels of these data are organized in another folder that contains 16 label files with the naming format of ‘Subxx_xxx_Label.csv’.
Technical Validation
Repeatability
Data repeatability often reflects the consistency and accuracy with which the subjects performed the movements. This phenomenon would normally influence data analysis outcomes in the case of pattern recognition and generalization of the data characteristics. Therefore, it is essential to investigate the data repeatability of the acquired dataset using multiple measures. Thus, the data repeatability has been examined and validated across subjects and evaluations metrics in a stepwise procedure as follows. Firstly, the active processes (STDUP, SITDN, KLFT, TPTO, LLF, LLB, LLS, KLCL, HS, TO, LUGF, and LUGB) of each subject were extracted from their individual data files. Secondly, the average angle of hip adduction, hip flexion, knee flexion, and ankle flexion in the left leg was calculated and stored. Thirdly, the average coefficient of determination between the mean angle and the joint angle was calculated. And finally, the R-square scores of each movement per subject were obtained and the average R-square values of the four joint angles were weighted to arrival at the final value, \(\overline{{R}^{2}}\). The weights corresponding to the four joint angles were then calculated according to the relative size of their motion ranges in each movement. This final value, \(\overline{{R}^{2}}\) represent a valid means for assessing the data repeatability of each movement per subject. The statistical bar plots (Fig. 6a) show that the \(\overline{{R}^{2}}\) values for the twelve movements (excluding WAK, UPS, DNS, and STC), most of them above 0.80, indicating good repeatability. The circles in Fig. 6a show the distribution of \({R}^{2}\) values of 40 subjects for twelve movements.
Analyses of the data repeatability and synchronisation. (a) The cycles show the average coefficient of determination (\(\overline{{R}^{2}}\)) between the average angle and the joint angle of each movement for each subject. The bar plots show the average \(\overline{{R}^{2}}\) of each movement. (b) The average gait cycle of walking on level ground, upstairs, and downstairs. (c) The synchronisation between the sEMG acquisition system and motion capture system.
Moreover, the gait cycle data of WAK, UPS, and DNS were extracted for calculating the average results of hip angle, knee angle, ankle angle, hip torque, knee torque, and ankle torque (as shown in Fig. 6b). Compared with other researchers’ results, the data in this work are most similar to the result of J. Camargo et al.16. They recorded WAK, UPS, and DNS with different speeds in their dataset, and the WAK data between slow speed and normal speed is closer to our result than fast speed, which reflects the average walking speed in our proposed dataset (SIAT-LLMD). At the same time, we also noticed some differences: our data on the minimum ankle angle is bigger than the results of J. Camargo et al.16, but smaller than E. Reznick et al.28. For the UPS, our results are close to the results between fast speed and normal speed in the work of J. Camargo et al.16 but still have some differences. Our minimum ankle angle in DNS is bigger than their results but similar to E. Reznick et al.28. Also, our maximum hip angle in DNS is about 7 degrees smaller than J. Camargo et al.16 and much smaller than the other datasets (the difference between these datasets about this value is also more than 20 degrees)14,28,29. The minimum and maximum hip angle in UPS of these datasets are different (the results of SIAT-LLMD are smallest), but the angular range in all these datasets are around 50 degrees14,16,28,29. For STDUP and SITDN, the kinematic and kinetic data can be mutually verified with the research of C. Pinheiro et al.30. However, it should be noted that due to the limited number of steps in the staircase, steady stair ascent or descent may not be achievable, which is a potential limitation of this dataset. Another limitation of this dataset is that the movements were chosen to achieve the above goals as much as possible, but it is important to note that the subjects without any special training may not be able to reach the same levels of performance as highly trained individuals due to limitations in their physical fitness and balance ability.
Synchronization
During the data collection, the experiment and equipment were set up in a manner that ensured the synchronization of the different data (sEMG recordings and data from the Motion Capture System). Figure 6c demonstrates the normalized knee angle and the normalized sEMG signal along the same time axis, where the angle changes rapidly with the strength of the sEMG signal, indicating the synchronization.
Classification and regression
To assess the extent to which the various classes of lower limb movements could be decoded, a support vector machine (SVM) classifier (that employs a polynomial Kernel function of order 2, ‘one vs one’ method) and a k-nearest neighbour (KNN) classifier (with K = 5) were applied to classify the 12 movements; a the same SVM and KNN (with K = 1) classifiers were utilized to classify the different gait periods during WAK, UPS, and DNS. In addition, a Gaussian process regression model was employed to predict the angle and torque associated with the joints during WAK, UPS, and DNS. The classification and regression performances were individually evaluated for each subject. Before building the classification and regression models, the sEMG data went through a sequence of filtering operation via filters that includes: notch (with a frequency: 50 Hz; filter quality factor: 100), Butterworth (with low cut-off frequency: 15 Hz; high cut-off frequency: 400 Hz; sampling frequency, fs: 1920 Hz, and order: 7), and wavelet and packet (with wavelet packet threshold: 0.08; wave packet threshold setting: soft; wavelet packet type: ‘db7’; level: 9). Subsequently, the filtered data was segmented using a sliding window (window size: 150 sample points for the classification of 12 movements; window size: 80 sample points for the classification of gait phase) and the Du’s sEMG feature set31 (include Integrated EMG, variance of EMG, waveform length, zero crossing, slope sign change, and Willison amplitude) was extracted. It should be noted that the feature set was normalized by using Min-Max scaling normalization method, and subsequently used for the characterization of the movement classes.
With the SVM classifier, the classification results show that the average accuracy of the 12 movements is 90.74%, while the WAK gait phase classification achieved an accuracy of 84.50%%, UPS gait phase recorded an accuracy of 88.77%, and DNS gait phase had an accuracy of 90.38%. Meanwhile, the KNN classifier yielded classification results with average accuracy of 85.06%%, across the 12 movement classes while the gait phase average classification results are 80.62% for WAK, 86.82% for UPS, and 89.78% for DNS. The results have been detailed in Figs. 7a, 8a with statistical bar plots indicating that the average accuracy of each movement or gait phase is much above the random selection probability. The error bars (Figs. 7a, 8a) reveal the distribution of the accuracy of each movement from the 40 subjects, presenting acceptable variance. Furthermore, the confusion matrix (Figs. 7b, 8b) shows that the movement with a lower accuracy has other movements similar to it, thereby confirming the reasonability of the results. To train and test our model, we utilized the 5-fold cross-validation technique provided by MATLAB’s Statistics and Machine Learning Toolbox.
For the regression analysis, the statistical bar plots (Fig. 9a) show the average Root Mean Square Error (RMSE) of each joint (the lower, the better), while the error bar (Fig. 9a) represents the distribution of RMSE of each joint-angle and joint-torque from the 40 subjects (the lower, the better). These errors are much smaller than their usual range as reported in a previous study32, indicating that the sEMG signal can effectively aid the prediction of the angle and torque associated with the joints. In addition, Fig. 9b shows the regression effect to be more intuitively, with the predicted and actual values presenting the same trend. In conclusion, the above results have reiterated the applicability of our dataset (SIAT-LLMD) from a multi-analysis perspective for lower limb movement intent recognition and gait phase characterization.
Code availability
A set of codes for reading, pre-processing of sEMG, splitting of sEMG into windows of various sizes, extracting of sEMG features (including 20 kinds of features33 that can be combined into Du’s feature set31, Hudgins’s feature set34, and novel time-domain feature set35,36), normalization of extracted features, generation of sample data, and making log files are provided for easy handling of the data. In addition, some programs in the technical validation section were also included, which can be found from on GITHUB via the following URL: https://github.com/WH-Wei/SIAT-Lower-Limb-Motion-Dataset-Codes.git.
References
Garro, F., Chiappalone, M., Buccelli, S., De Michieli, L. & Semprini, M. Neuromechanical Biomarkers for Robotic Neurorehabilitation. Front. Neurorobot. 15, (2021).
Fleming, A. et al. Myoelectric control of robotic lower limb prostheses: A review of electromyography interfaces, control paradigms, challenges and future directions. J. Neural Eng. 18, (2021).
Li, W., Shi, P. & Yu, H. Gesture Recognition Using Surface Electromyography and Deep Learning for Prostheses Hand: State-of-the-Art, Challenges, and Future. Front. Neurosci. 15, (2021).
Samuel, O. W. et al. Resolving the adverse impact of mobility on myoelectric pattern recognition in upper-limb multifunctional prostheses. Comput. Biol. Med. 90, 76–87 (2017).
Samuel, O. W. et al. Intelligent EMG pattern recognition control method for upper-limb multifunctional prostheses: Advances, current challenges, and future prospects. IEEE Access 7, 10150–10165 (2019).
Ali, A., Fontanari, V., Schmoelz, W. & Agrawal, S. K. Systematic Review of Back-Support Exoskeletons and Soft Robotic Suits. Front. Bioeng. Biotechnol. 9, 1–15 (2021).
Fleischer, C., Reinicke, C. & Hommel, G. Predicting the intended motion with EMG signals for an exoskeleton orthosis controller. 2005 IEEE/RSJ Int. Conf. Intell. Robot. Syst. IROS 2029–2034, https://doi.org/10.1109/IROS.2005.1545504 (2005).
Xu, H. & Xiong, A. Advances and disturbances in sEMG-Based intentions and movements recognition: A review. IEEE Sens. J. 21, 13019–13028 (2021).
Atzori, M. et al. Characterization of a benchmark database for myoelectric movement classification. IEEE Trans. Neural Syst. Rehabil. Eng. 23, 73–83 (2015).
Atzori, M. et al. Electromyography data for non-invasive naturally-controlled robotic hand prostheses. Sci. Data 1, 1–13 (2014).
Schreiber, C. & Moissenet, F. A multimodal dataset of human gait at different walking speeds established on injury-free adult participants. Sci. Data 6, 111 (2019).
Lencioni, T., Carpinella, I., Rabuffetti, M., Marzegan, A. & Ferrarin, M. Human kinematic, kinetic and EMG data during different walking and stair ascending and descending tasks. Sci. Data 6, 309 (2019).
Moreira, L., Figueiredo, J., Fonseca, P., Vilas-Boas, J. P. & Santos, C. P. Lower limb kinematic, kinetic, and EMG data from young healthy humans during walking at controlled speeds. Sci. Data 8, 1–11 (2021).
Luan, Y. et al. HAR-sEMG: A Dataset for Human Activity Recognition on Lower-Limb sEMG. Knowl. Inf. Syst. 63, 2791–2814 (2021).
Hu, B., Rouse, E. & Hargrove, L. Benchmark Datasets for Bilateral Lower-Limb Neuromechanical Signals from Wearable Sensors during Unassisted Locomotion in Able-Bodied Individuals. Front. Robot. AI 5, 1–5 (2018).
Camargo, J., Ramanathan, A., Flanagan, W. & Young, A. A comprehensive, open-source dataset of lower limb biomechanics in multiple conditions of stairs, ramps, and level-ground ambulation and transitions. J. Biomech. 119, 110320 (2021).
Wojtusch, J. & Von Stryk, O. HuMoD - A versatile and open database for the investigation, modeling and simulation of human motion dynamics on actuation level. IEEE-RAS Int. Conf. Humanoid Robot. 2015-Decem, 74–79 (2015).
Wei, M., Liu, Q., Zhou, Z. & Ai, Q. Active interaction control applied to a lower limb rehabilitation robot by using EMG recognition and impedance model. Ind. Rob. 41, 465–479 (2014).
Lendaro, E., Mastinu, E., Håkansson, B. & Ortiz-Catalan, M. Real-time classification of non-weight bearing lower-limb movements using EMG to facilitate phantom motor execution: Engineering and case study application on phantom limb pain. Front. Neurol. 8, 1–12 (2017).
Wei, W. et al. Surface Electromyogram, Kinematic, and Kinetic Dataset of Lower Limb Walking for Movement Intent Recognition, figshare, https://doi.org/10.6084/m9.figshare.c.6271164.v1 (2023).
John, C. T., Anderson, F. C., Higginson, J. S. & Delp, S. L. Stabilisation of walking by intrinsic muscle properties revealed in a three-dimensional muscle-driven simulation. Comput. Methods Biomech. Biomed. Engin. 16, 451–462 (2013).
Wu, G. et al. ISB recommendation on definitions of joint coordinate system of various joints for the reporting of human joint motion—part I: ankle, hip, and spine. J. Biomech. 35, 543–548 (2002).
Wu, G. et al. ISB recommendation on definitions of joint coordinate systems of various joints for the reporting of human joint motion—Part II: shoulder, elbow, wrist and hand. J. Biomech. 38, 981–992 (2005).
Stegeman, D. & Hermens, H. Standards for surface electromyography: The European project Surface EMG for non-invasive assessment of muscles (SENIAM). Línea). Dispon. en http//www.med.… 108–112 (2007).
Martinez-Villalpando, E. C. & Herr, H. Agonist-antagonist active knee prosthesis: A preliminary study in level-ground walking. J. Rehabil. Res. Dev. 46, 361–373 (2009).
Rouse, E. J., Mooney, L. M. & Herr, H. M. Clutchable series-elastic actuator: Implications for prosthetic knee design. Int. J. Rob. Res. 33, 1611–1625 (2014).
Wang, D., Liu, M., Zhang, F. & Huang, H. Design of an expert system to automatically calibrate impedance control for powered knee prostheses. in 2013 IEEE 13th International Conference on Rehabilitation Robotics (ICORR) 1–5 (IEEE, 2013).
Reznick, E. et al. Lower-limb kinematics and kinetics during continuously varying human locomotion. Sci. Data 8, 1–12 (2021).
Fukuchi, C. A., Fukuchi, R. K. & Duarte, M. A public dataset of overground and treadmill walking kinematics and kinetics in healthy individuals. PeerJ 2018, 1–17 (2018).
Pinheiro, C. et al. Kinematic and kinetic study of sit-to-stand and stand-to-sit movements towards a human-like skeletal model. in 2019 IEEE 6th Portuguese Meeting on Bioengineering (ENBENG) 1–4, https://doi.org/10.1109/ENBENG.2019.8692569 (IEEE, 2019).
Du, Y. C., Shyu, L. Y. & Hu, W. The effect of combining stationary wavelet transform and independent component analysis in the multichannel SEMGs hand motion identification system. Journal of Medical and Biological Engineering vol. 26 9–14 at (2006).
Riener, R., Rabuffetti, M. & Frigo, C. Stair ascent and descent at different inclinations. Gait Posture 15, 32–44 (2002).
Phinyomark, A., Phukpattaranont, P. & Limsakul, C. Feature reduction and selection for EMG signal classification. Expert Syst. Appl. 39, 7420–7431 (2012).
Hudgins, B., Parker, P. & Scott, R. N. A new strategy for multifunction myoelectric control. IEEE Trans. Biomed. Eng. 40, 82–94 (1993).
Samuel, O. W. et al. A Novel Time-Domain Descriptor for Improved Prediction of Upper Limb Movement Intent in EMG-PR System. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS 2018(July), 3513–3516 (2018).
Samuel, O. W. et al. Pattern recognition of electromyography signals based on novel time domain features for amputees’ limb motion classification. Comput. Electr. Eng. 67, 646–655 (2018).
Toledo-Pérez, D. C., Martínez-Prado, M. A., Gómez-Loenzo, R. A., Paredes-García, W. J. & Rodríguez-Reséndiz, J. A study of movement classification of the lower limb based on up to 4-EMG channels. Electron. 8, 1–11 (2019).
Wang, J., Cao, D., Wang, J. & Liu, C. Action Recognition of Lower Limbs Based on Surface Electromyography Weighted Feature Method. Sensors 21, 6147 (2021).
Wang, J., Dai, Y. & Si, X. Analysis and Recognition of Human Lower Limb Motions Based on Electromyography (EMG) Signals. Electronics 10, 2473 (2021).
Zhou, C., Yang, L., Liao, H., Liang, B. & Ye, X. Ankle foot motion recognition based on wireless wearable sEMG and acceleration sensors for smart AFO. Sensors Actuators A Phys. 331, 113025 (2021).
Peng, Z., Junxia, Z. & Elsabba, A. M. M. Lower Limb Motion Intention Recognition Based on sEMG Fusion Features. IEEE Sens. J. XX, (2022).
Li, X., Tian, L., Zheng, Y. & Li, G. Real-time performance test evaluation system for lower limb motion intention recognition algorithm. Chinese J. Sci. Instrum. 41, 99–107 (2020).
Xin, S., Zhirui, F., Jieyi, Z., Shuyuan, X. & Jianning, C. Rapid classification of lower limb movements of EMG signals based on LMS-random forest. Chinese J. Sci. Instrum. 41, 218 (2020).
Wang, X. et al. sEMG-based consecutive estimation of human lower limb movement by using multi-branch neural network. Biomed. Signal Process. Control 68, 102781 (2021).
Yuan, Y. et al. Gait Phase Classification Based on sEMG Signals Using Long Short-Term Memory for Lower Limb Exoskeleton Robot. IOP Conf. Ser. Mater. Sci. Eng. 853, 012041 (2020).
Ryu, J., Lee, B. H. & Kim, D. H. sEMG signal-based lower limb human motion detection using a top and slope feature extraction algorithm. IEEE Signal Process. Lett. 24, 929–932 (2017).
Joshi, C. D., Lahiri, U. & Thakor, N. V. Classification of Gait Phases from Lower Limb EMG: Application to Exoskeleton Orthosis. 2013 Ieee Point-of-Care Healthc. Technol. 228–231 (2013).
Chen, J., Zhang, X., Cheng, Y. & Xi, N. Surface EMG based continuous estimation of human lower limb joint angles by using deep belief networks. Biomed. Signal Process. Control 40, 335–342 (2018).
Mamikoglu, U. et al. Electromyography based joint angle estimation and control of a robotic leg. in 2016 6th IEEE International Conference on Biomedical Robotics and Biomechatronics (BioRob) 182–187 (IEEE, 2016).
Li, Q. L., Song, Y. & Hou, Z. G. Estimation of Lower Limb Periodic Motions from sEMG Using Least Squares Support Vector Regression. Neural Process. Lett. 41, 371–388 (2015).
Zhang, F. et al. sEMG-based continuous estimation of joint angles of human legs by using BP neural network. Neurocomputing 78, 139–148 (2012).
Khanjani, I., Khoshdel, V. & Akbarzadeh, A. Estimate human-force from sEMG signals for a lower-limb rehabilitation robot. in 2017 Iranian Conference on Electrical Engineering (ICEE) 132–136, https://doi.org/10.1109/IranianCEE.2017.7985275 (IEEE, 2017).
Peng, L. et al. sEMG-based torque estimation for robot-assisted lower limb rehabilitation. in 2015 International Joint Conference on Neural Networks (IJCNN) vols 2015-Septe 1–5 (IEEE, 2015).
Di Nardo, F., Mengarelli, A., Maranesi, E., Burattini, L. & Fioretti, S. Gender differences in the myoelectric activity of lower limb muscles in young healthy subjects during walking. Biomed. Signal Process. Control 19, 14–22 (2015).
Fukuchi, R. K., Fukuchi, C. A. & Duarte, M. A public dataset of running biomechanics and the effects of running speed on lower extremity kinematics and kinetics. PeerJ 2017, 3298 (2017).
Acknowledgements
This project is in part supported by the grants from the National Natural Science Foundation of China (#U1913601), the National Key Research and Development Program of China (#2018YFA0701405), Guangdong Basic and Applied Research Foundation (#2023A1515011478), and Guangdong-Hong Kong-Macao Joint Laboratory of Human-Machine Intelligence-Synergy Systems.
Author information
Authors and Affiliations
Contributions
G.L. conceived the study; W.W. designed and performed experiments, processed and analysed the data; F.T. performed experiments, processed the data, made labels, and checked the correctness of the data; W.W., O.W.S., G.L. interpreted the data, wrote the manuscript and revised it at all the stages of the project. H.Z. designed and performed experiments; H.M. performed the experiments with the female subject; M.L. optimized the data presentation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wei, W., Tan, F., Zhang, H. et al. Surface electromyogram, kinematic, and kinetic dataset of lower limb walking for movement intent recognition. Sci Data 10, 358 (2023). https://doi.org/10.1038/s41597-023-02263-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02263-3
- Springer Nature Limited