Abstract
Computer-aided diagnosis systems in adult chest radiography (CXR) have recently achieved great success thanks to the availability of large-scale, annotated datasets and the advent of high-performance supervised learning algorithms. However, the development of diagnostic models for detecting and diagnosing pediatric diseases in CXR scans is undertaken due to the lack of high-quality physician-annotated datasets. To overcome this challenge, we introduce and release PediCXR, a new pediatric CXR dataset of 9,125 studies retrospectively collected from a major pediatric hospital in Vietnam between 2020 and 2021. Each scan was manually annotated by a pediatric radiologist with more than ten years of experience. The dataset was labeled for the presence of 36 critical findings and 15 diseases. In particular, each abnormal finding was identified via a rectangle bounding box on the image. To the best of our knowledge, this is the first and largest pediatric CXR dataset containing lesion-level annotations and image-level labels for the detection of multiple findings and diseases. For algorithm development, the dataset was divided into a training set of 7,728 and a test set of 1,397. To encourage new advances in pediatric CXR interpretation using data-driven approaches, we provide a detailed description of the PediCXR data sample and make the dataset publicly available on https://physionet.org/content/vindr-pcxr/1.0.0/.
Similar content being viewed by others
Background & Summary
Common thoracic diseases cause several hundred thousand deaths every year among children under five years old1,2. The chest radiograph or CXR is the first-line and most commonly performed imaging examination in the assessment of the pediatric patient3. Interpreting CXR scans on pediatric patients can be for a number of indications or critical findings, in particular for common thoracic diseases in children such as Pneumonia, Bronchitis and Cardiovascular diseases (CVDs). Depending on the patients’ age, the difficulty of the examination will vary, often requiring a specialist in pediatric diagnostic imaging with an in-depth knowledge of radiological signs of different lung conditions4. Additionally, the inter-observer agreement and intra-observer agreement in the pediatric CXR interpretation were low5. This opens room for the development of data-driven approaches and computational tools to assist pediatricians in the diagnosis of common thoracic diseases and to reduce their workload.
Computer-aided diagnosis (CAD) systems for identification of lung abnormality in adult CXRs have recently achieved great success thanks to the availability of large labeled datasets6,7,8,9,10. Many large-scale CXR datasets of adult patients such as Montgomery County chest X-ray (MC)11, Shenzhen chest X-ray11, ChestX-ray86, COVIDGR12, ChestX-ray146, Padchest7, CheXpert8, MIMIC-CXR9 and VinDr-CXR10 have been established and released in recent years. These datasets boosted new advances in exploring new machine learning-based approaches in the interpretation of CXR in adults8,13,14,15,16,17,18. Unfortunately, the creation of pediatric CXR datasets is still unexploited, and the number of benchmark pediatric CXR datasets is limited. This becomes the main obstacle in developing and transferring new machine learning-based CAD systems for pediatric CXR in clinical practice.
In an effort to provide a large-scale pediatric CXR dataset with high-quality annotations for the research community, we have built the PediCXR dataset in DICOM format. The dataset consists of 9,125 posteroanterior (PA) view CXR scans in patients younger than 10 years that were retrospectively collected from three major hospitals in Vietnam from 2020 to 2021. In particular, all CXR scans come with both the localization of critical findings and the classification of common thoracic diseases. These images were annotated by a group of three radiologists with at least 10 years of experience for the presence of 36 critical findings (local labels) and 15 diagnoses (global labels). Here, the local labels should be annotated with rectangle bounding boxes that localize the findings, while the global labels reflect the diagnostic impression of the radiologist at the image-level. For algorithm development, we randomly divided the dataset into two parts: the training set of 7,728 scans (84.7%) and the test set of 1,397 scans (15.3%). To the best of our knowledge, the released PediCXR is currently the largest public pediatric CXR dataset with radiologist-generated annotations in both training and test sets. Table 1 below shows an overview of existing public datasets for CXR interpretation in pediatric patients, compared with the PediCXR. Compared to the previous works, the PediCXR dataset shows two main advantages. First, the dataset is labeled for multiple findings and diseases. Meanwhile, most pediatric CXR datasets have focused on a single disease such as pneumonia19 or pneumothorax20. Second, the dataset provides bounding box annotations at lesion level, which is useful for developing explainable artificial intelligent models21 for the CXR interpretation in children. We believe the introduction of the PediCXR provides a suitable imaging source for investigating the ability of supervised machine learning models in identifying common lung diseases in pediatric patients.
Methods
Data collection
Data collection was conducted at the Phu Tho Obstetric & Pediatric Hospital (PTOPH) between 2020–2021. The ethical clearance of this study was approved by the Institutional Review Boards (IRBs) of the PTOPH. The need for obtaining informed patient consent was waived because this retrospective study did not impact clinical care or workflow at these two hospitals, and all patient-identifiable information in the data has been removed. We retrospectively collected more than 10,000 CXRs in DICOM format from a local picture archiving and communication system (PACS) at PTOPH. The imaging dataset was then transferred and analyzed at Smart Health Center, VinBigData JSC.
Overview of approach
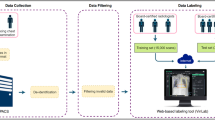
The building of the PediCXR dataset is illustrated in Fig. 1. In particular, the collection and normalization of the dataset were divided into four main steps: (1) data collection, (2) data de-identification, (3) data filtering, and (4) data labeling. We describe each step in detail as below.
Construction of the PediCXR dataset. First, raw pediatric scans in DICOM format were collected retrospectively from the hospital’s PACS at PTOPH. These images were de-identified to protect patient’s privacy. Then, invalid files (including adult CXR images, images of other modalities or other body parts, images with low quality, or incorrect orientation) were manually filtered out. After that, a web-based DICOM labeling tool called VinDr Lab was developed to remotely annotate DICOM data. Finally, the annotated dataset was then divided into a training set (N = 7,728) and a test set (N = 1,397) for algorithm development.
Data de-identification
In this study, we follow the HIPAA Privacy Rule22 to protect individually identifiable health information from the DICOM images. To this end, we removed or replaced with random values all personally identifiable information associated with the images via a two-stage de-identification process. At the first stage, a Python script was used to remove all DICOM tags of protected health information (PHI)23 such as patient’s name, patient’s date of birth, patient ID, or acquisition time and date, etc. For the purpose of loading and processing DICOM files, we only retained a limited number of DICOM attributes that are necessary, as indicated in Table 2 (Supplementary materials). In the second stage, we manually removed all textual information appearing on the image data, i.e., pixel annotations that could include patient’s identifiable information.
Data filtering
The collected raw data included a significant amount of outliers including CXRs of adult patients, body parts other than chest (abdominal, spine, and others), low-quality images, or lateral CXRs. To filter a large number of CXR scans, we trained a lightweight convolutional neural network (CNN)24 to remove all outliers automatically. Next, a manual verification was performed to ensure all outliers had been fully removed.
Data labeling
The PediCXR dataset was labeled for a total of 36 findings and 15 diagnoses. These labels were divided into two categories: local labels (#1– #36) and global labels (#37– #52). The local labels should be marked with bounding boxes that localize the findings, while the global labels should reflect the diagnostic impression of the radiologist. This list of labels was suggested by a committee of the most experienced pediatric radiologists. To select these labels, the committee took into account two key factors. First, findings and diseases are prevalent. Second, they can be differentiated on pediatric chest X-ray scans. Figure 2 illustrates several samples with both local and global labels annotated by our radiologists.
Several examples of pediatric CXR images with radiologist’s annotations. Local labels marked by radiologists are plotted on the original images for visualization purposes. These annotations show abnormal findings from the scans. The global labels, that classify images into diseases, are in bold and listed at the bottom of each example.
To facilitate the labeling process, we designed and built a web-based framework called VinDr Lab25 that allows a team of experienced radiologists remotely annotate the data. Specifically, this is a web-based labeling tool that was developed to store, manage, and remotely annotate DICOM data. The radiologists were oriented to locate the abnormal findings from the DICOM viewer and draw the bounding boxes. All the annotators have been well-trained to ensure that the annotations are consistently annotated. In addition, all the radiologists participating in the labeling process were certified in diagnostic radiology and received healthcare professional certificates. In total, three pediatric radiologists with at least 15 years of experience were involved in the annotation process. Each sample in the training set was assigned to one radiologist for annotation. Additionally, all of the participating radiologists were blinded to relevant clinical information. A set of 9,125 pediatric CXRs were randomly annotated from the filtered data, of which 7,728 scans serve as the training set, and the remaining 1,397 studies form the test set. Note the 9,125 studies correspond to 9,125 patients, and each study has a single CXR scan.
Once the labeling was completed, the annotations of all pediatric CXRs were exported in JavaScript Object Notation (JSON) format. We developed a Python script to parse JSON files and organized the annotations in the form of a single comma-separated values (CSV) file. Each CSV file contains labels, bounding box coordinates, and their corresponding image identifiers (IDs). The data characteristics, including patient demographic and the prevalence of each finding or disease, are summarized in Table 3. The distributions of abnormal findings and pathologies in the training set are drawn in Figs. 3, 4, respectively.
Data Records
The PediCXR dataset will be made available for public download on PhysioNet26. We offer complete imaging data as well as ground truth labels for both the training and test datasets. The pediatric scans were split into two folders: one for training and one for testing, named as “train” and “test”, respectively. Since each study has only one instance and each patient has maximum one study, therefore, the value of the SOP Instance UID provided by the DICOM tag (0008,0018) was encoded into a unique, anonymous identifier for each image. To this end, we used the Python hashlib module (see Code Availability) to encode the SOP Instance UIDs into image IDs. The radiologists’ local annotations of the training set were provided in a CSV file called annotations_train.csv. Each row of the CSV file represents a bounding box annotation with the following attributes: image ID (image_id), radiologist ID (rad_id), label’s name (class_name), bounding box coordinates (x_min, y_min, x_max, y_max), and label class ID (class_id). The coordinates of the box’s upper-left corner are (x_min, y_min), and the coordinates of the box’s lower right corner are (x_max, y_max). Meanwhile, the image-level labels of the training set were stored in a different CSV file called image_labels_train.csv, with the following fields: Image ID (image_id), radiologist ID (rad_ID), and labels (labels) for both the findings and diagnoses. Each image ID is associated with a vector of multiple labels corresponding to different pathologies, with positive pathologies encoded as “1” and negative pathologies encoded as “0”. Similarly, the test set’s bounding-box annotations and image-level labels were saved in the files annotations_test.csv and image_labels_test.csv, respectively.
Technical Validation
The data de-identification process was controlled. Specifically, all DICOM meta-data was parsed and manually reviewed to ensure that all individually identifiable health information (PHI)23 of the children patients has been removed to meet the U.S. HIPAA22 regulations. In addition, pixel values of all pediatric CXR scans were also carefully examined by human readers. During this review process, all scans were manually reviewed case-by-case by a team of 10 human readers. A small number of images containing private textual information that had not been removed by our algorithm was excluded from the dataset. The manual review process also helped identify and discard out-of-distribution samples such as CXRs of adult patients, body parts other than the chest, low-quality images, or lateral CXRs that our machine learning classifier was not able to detect. A set of rules underlying our web-based annotation tool were developed to control the quality of the labeling process. These rules prevent human annotators from mechanical mistakes like forgetting to choose global labels or marking lesions on the image while choosing “No finding” as the global label.
Usage Notes
The PediCXR dataset was established for the purpose of developing and evaluating machine learning algorithms for detecting and localizing anomalies in pediatric CXR images. The dataset has been previously used in a study on the diagnosis of multiple diseases in pediatric patients27 and showed promising results. Specifically, the authors27 introduced a deep learning network to detect common pulmonary pathologies on CXR of pediatric patients. On the test set of 777 studies of the PediCXR dataset, the network yielded an area under the receiver operating characteristic (AUC) of 0.709 (95% CI, 0.690–0.729). The sensitivity, specificity, and F1-score at the cutoff value are 0.722 (0.694–0.750), 0.579 (0.563–0.595), and 0.389 (0.373–0.405), respectively. However, they recognized that its performance remains low compared to medical experts. This work revealed the major challenge in learning disease features on pediatric CXR images using representation learning techniques, opening huge aspects for future research.
The primary uses for which the PediCXR dataset was conceptualized include:
-
Developing and validating a predictive model for the classification of common thoracic diseases in pediatric patients.
-
Developing and validating a predictive model for the localization of multiple abnormal findings on the pediatric chest X-ray scans.
Finally, the released dataset remains with limitations that still need to be addressed in the future, including:
-
The dataset did not contain clinical information associated with DICOM images, which is essential for the interpretation of CXR in children patients.
-
The number of examples for rare diseases (e.g., Congenital pulmonary airway malformation (CPAM), Congenital emphysema, Diagphramatic hernia, Mediastinal tumor, Pleuro-pneumonia, Situs inversus, Lung tumor) or findings (Emphysema, Edema, Calcification, Chest wall mass, Bronchectasis, Pleural thickening, Clavicle fracture, Pleuropulmonary mass, Paraveterbral mass, etc.) are limited. Hence, training supervised learning algorithms, which requires a large-scale annotated dataset, on the PediCXR dataset to diagnose the rare diseases and findings is not reliable.
To download and use the PediCXR, users are required to accept the https://physionet.org/content/mimic-cxr/view-license/2.0.0/PhysioNet Credentialed Health Data License 1.5.0. By accepting this license, users agree that they will not share access to the dataset with anyone else. For any publication that explores this resource, the authors must cite this original paper and release their code and models.
Code availability
This study used the following open-source repositories to load and process DICOM scans: Python 3.7.0 (https://www.python.org/); Pydicom 1.2.0 (https://pydicom.github.io/); OpenCV-Python 4.2.0.34 (https://pypi.org/project/opencv-python/); and Python hashlib (https://docs.python.org/3/library/hashlib.html). The code for data de-identification was made publicly available at https://github.com/vinbigdata-medical/vindr-cxr. The code to train CNN classifier for the out-of-distribution task was made publicly available at https://github.com/vinbigdata-medical/DICOM-Imaging-Router. The VinDr Lab is an open source software and can be found at https://vindr.ai/vindr-lab.
References
Collaborators, G. L. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the global burden of disease study 2015. The Lancet Infect. Dis. 17, 1133–1161 (2017).
Wardlaw, T. M., Johansson, E. W., Hodge, M., Organization, W. H. & (UNICEF), U. N. C. F. Pneumonia: The forgotten killer of children (2006).
Hart, A. & Lee, E. Y. Pediatric Chest Disorders: Practical Imaging Approach to Diagnosis. Dis. Chest, Breast, Hear. Vessel. 2019-2022 107–125 (2019).
Chest radiograph (pediatric). https://radiopaedia.org/articles/chest-radiograph-paediatric. Accessed: 2021-09-24.
Du Toit, G., Swingler, G. & Iloni, K. Observer variation in detecting lymphadenopathy on chest radiography. Int. J. Tuberc. Lung Dis. 6, 814–817 (2002).
Wang, X. et al. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2097–2106, https://doi.org/10.1109/CVPR.2017.369 (2017).
Bustos, A., Pertusa, A., Salinas, J.-M. & de la Iglesia-Vayá, M. Padchest: A large chest X-ray image dataset with multi-label annotated reports. arXiv preprint arXiv:1901.07441 (2019).
Irvin, J. et al. CheXpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence 33, 590–597 (2019).
Johnson, A. E. et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 6, 317, https://doi.org/10.1038/s41597-019-0322-0 (2019).
Nguyen, H. Q. et al. VinDr-CXR: An open dataset of chest X-rays with radiologist’s annotations. Sci. Data 9, 429 (2022).
Jaeger, S. et al. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Medicine Surg. 4, 475–477, https://doi.org/10.3978/j.issn.2223-4292.2014.11.20 (2014).
Tabik, S. et al. COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest X-ray images. IEEE journal biomedical health informatics 24, 3595–3605 (2020).
Rajpurkar, P. et al. CheXNet: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprint arXiv:1711.05225 (2017).
Rajpurkar, P. et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Medicine 15, e1002686, https://doi.org/10.1371/journal.pmed.1002686 (2018).
Majkowska, A. et al. Chest radiograph interpretation with deep learning models: Assessment with radiologist adjudicated reference standards and population-adjusted evaluation. Radiology 294, 421–431, https://doi.org/10.1148/radiol.2019191293 (2020).
Rajpurkar, P. et al. CheXpedition: Investigating generalization challenges for translation of chest X-ray algorithms to the clinical setting. arXiv preprint arXiv:2002.11379 (2020).
Tang, Y.-X. et al. Automated abnormality classification of chest radiographs using deep convolutional neural networks. npj Digit. Medicine 3, 1–8, https://doi.org/10.1038/s41746-020-0273-z (2020).
Pham, H. H., Le, T. T., Tran, D. Q., Ngo, D. T. & Nguyen, H. Q. Interpreting chest X-rays via CNNs that exploit hierarchical disease dependencies and uncertainty labels. Neurocomputing 437, 186–194 (2021).
Kermany, D. S. et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 172, 1122–1131.e9, https://doi.org/10.1016/j.cell.2018.02.010 (2018).
Chen, K.-C. et al. Diagnosis of common pulmonary diseases in children by X-ray images and deep learning. Sci. Reports 10, 1–9 (2020).
Gordon, L., Grantcharov, T. & Rudzicz, F. Explainable artificial intelligence for safe intraoperative decision support. JAMA surgery 154, 1064–1065 (2019).
US Department of Health and Human Services. Summary of the HIPAA privacy rule. https://www.hhs.gov/hipaa/for-professionals/privacy/laws-regulations/index.html (2003).
Isola, S. & Al Khalili, Y. Protected Health Information (PHI). https://www.ncbi.nlm.nih.gov/books/NBK553131/ (2019).
Pham, H. H., Do, D. V. & Nguyen, H. Q. DICOM Imaging Router: An Open Deep Learning Framework for Classification of Body Parts from DICOM X-ray Scans. arXiv preprint arXiv:2108.06490 (2021).
Nguyen, N. T. et al. VinDr Lab: A Data Platform for Medical AI. URL: https://github.com/vinbigdata-medical/vindr-lab (2021).
Pham, H. H., Tran, T. T. & Nguyen, H. Q. PediCXR: An open, large-scale pediatric chest X-ray dataset for interpretation of common thoracic diseases (version 1.0.0). PhysioNet https://doi.org/10.13026/k8qc-na36 (2022).
Tran, T. T. et al. Learning to automatically diagnose multiple diseases in pediatric chest radiographs using deep convolutional neural networks. In IEEE Conference on Computer Vision and Pattern Recognition Workshop (ICCV 2021) (2021).
Acknowledgements
The collection of this dataset was funded by the Smart Health Center, VinBigData JSC. The authors would like to acknowledge the Phu Tho Obstetric & Pediatric Hospital for agreeing to make the PediCXR dataset publicly available. We are especially thankful to Anh T. Nguyen, Huong T.T. Nguyen, Ngan T.T. Nguyen for their helps in the data collection and labeling process.
Author information
Authors and Affiliations
Contributions
H.Q.N. and H.H.P. designed the study; T.T.T. performed the data de-identification; H.Q.N. and H.H.P. wrote the paper; all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
This work was funded by the Vingroup JSC. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pham, H.H., Nguyen, N.H., Tran, T.T. et al. PediCXR: An open, large-scale chest radiograph dataset for interpretation of common thoracic diseases in children. Sci Data 10, 240 (2023). https://doi.org/10.1038/s41597-023-02102-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02102-5
- Springer Nature Limited