Abstract
Cold seep microbial communities are fascinating ecosystems on Earth which provide unique models for understanding the living strategies in deep-sea distinct environments. In this study, 23 metagenomes were generated from samples collected in the Site-F cold seep field in South China Sea, including the sea water closely above the invertebrate communities, the cold seep fluids, the fluids under the invertebrate communities and the sediment column around the seep vent. By binning tools, we retrieved a total of 768 metagenome assembled genome (MAGs) that were estimated to be >60% complete. Of the MAGs, 61 were estimated to be >90% complete, while an additional 105 were >80% complete. Phylogenomic analysis revealed 597 bacterial and 171 archaeal MAGs, of which nearly all were distantly related to known cultivated isolates. In the 768 MAGs, the abundant Bacteria in phylum level included Proteobacteria, Desulfobacterota, Bacteroidota, Patescibacteria and Chloroflexota, while the abundant Archaea included Asgardarchaeota, Thermoplasmatota, and Thermoproteota. These results provide a dataset available for further interrogation of deep-sea microbial ecology.
Measurement(s) | metagenome assembled genomes |
Technology Type(s) | metagenome sequencing and genome binning |
Sample Characteristic - Organism | microorganism |
Sample Characteristic - Environment | marine cold seep biome |
Sample Characteristic - Location | South China Sea |
Similar content being viewed by others
Background & Summary
Cold seeps are seafloor manifestations of methane-rich fluid migration from the sedimentary subsurface and support unique communities via chemosynthetic interactions fuelled1. The microorganisms inhabiting cold seeps transform the chemical energy in methane to products that sustain rich benthic communities around the gas leaks2. The use of next-generation sequencing methods has tremendously improved the insights into seep microbiomes and will advance microbial ecology from the diversity microbial distribution pattern to the adaptive survival strategy in deep-sea environments.
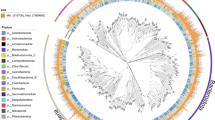
The cold seep in Site F (also known as Formosa Ridge) is one of the active cold seeps on the north-eastern slope of the South China Sea (SCS)3, where the natural gas hydrate exposed on the seafloor and was covered by chemosynthetic communities mainly comprising deep-sea mussels and galatheid crabs4. The geochemical characters have been illustrated by the in-situ detection using the developed Raman insertion Probe (RiP) system and integrated sensors5,6,7. The horizontal and vertical variations in methane concentrations showed contrasting trends in fields from the center of flourishing communities to the margin of sediments6. No CH4 or H2S Raman peaks were detected in the cold seep fluids, while dissolved CH4 were identified in the fluids under the lush chemosynthetic communities, and the sediment pore water profiles collected near the cold seep were characterized by the loss of SO42− and increased CH4, H2S and HS− peaks5,7. As the microbial communities in deep-sea cold seeps are often shaped by geochemical components in seepage solutions, we collected samples from the Site-F cold seep field in 2017, including the sea water closely above the invertebrate communities, the cold seep fluids, the fluids under the invertebrate communities and the sediment column around the seep vent (Fig. 1 and Table 1). The metagenomes were sequenced with Illumina HiSeq X Ten platform, with each metagenome yielding approximately 52.7 Gbps to 80.6 Gbps of clean bases (Table 2). We further obtained 768 metagenome-assembled genomes (MAGs) of environmental Bacteria and Archaea estimated to be >60% complete and <20% contamination (Supplementary Table 1). Of the MAGs, 61 were estimated to be >90% complete, while an additional 105 were >80% complete. There were 59 high-quality MAGs (completeness > 90% and contamination < 5%), accounting for 7.68% of the total. The anaerobic methanotrophic archaea (ANME), aerobic methanotrophic bacteria Methylococcales, sulfate-reducing Desulfobacterales, as well as sulfide-oxidizing Campylobacterales and Thiotrichales (Supplementary Table 2), well match the most favourable microbial metabolisms at methane seeps in terms of substrate supply. Meanwhile, the phylogenomic analysis suggests that this set of draft genomes includes highly sought-after genomes that lack cultured representatives, such as archaea Bathyarchaeota (30), Aenigmarchaeota (29), Heimdallarchaeota (20) and Pacearchaeota (10), and bacteria Patescibacteria (44), WOR-3 (23), Zixibacteria (13), Marinisomatota (12) and Eisenbacteria (6) et al. (Fig. 2). In addition, there are also some potential new phylum including NPL-UPA2 (7), UBP15 (4), FCPU426 (2) and SM23–31 (2) et al. All the non-redundant draft metagenome-assembled genomes described here were deposited into the National Center for Biotechnology Information (NCBI). These data will hopefully provide a resource for downstream analysis acting as references for largescale comparative genomics within globally vital phylogenetic groups, as well as allowing for the exploration of novel microbial metabolisms.
Sample collection and data analysis process. (a) Location and the sampling area in the cold seep field in the northern South China Sea. (b) Schematic overview of sampling and metagenomic analysis performed in this study. Each rectangle symbolizes processes containing descriptions (in bold), methods or tools used in the corresponding analysis.
Phylogenetic diversity of 768 metagenome assembled genomes (MAGs) from cold seep in South China Sea (Supplementary Table 2) and reference genomes of Bacteria and Archaea available in RefSeq (Supplementary Table 3). The scale bar corresponds to 3.00 substitutions per amino acid position. The number of draft genomes in each node are provided. The branches with red dots have no cultured representatives.
Methods
Sampling
Samples were retrieved from a cold seep field in the northern SCS by the KEXUE research vessel during the cruise in Sep 2017 (Fig. 1 a and Table 1). The water closely above the invertebrate communities was collected by an in-situ water sampling cylinder equipped on FAXIAN Remotely Operated Vehicle (ROV) during the dive 164 and 165 (sample ID: SW_1 and SW_2, respectively). The cold seep fluid was collected at the gas plumes during the dive 166 (sample ID: SW_3), and the fluid under the invertebrate communities was collected during the dive 167 (sample ID: SW_4). About 15 L water of each sample was filtered through a 0.22μm polycarbonate membrane (Millipore, Bedford, MA, USA). The membranes were stored at −80 °C and used for DNA extraction. A sediment core was collected by ROV at reductive sediments area nearby the invertebrate communities during dive 157. A thin outer layer ( < 1 cm) of the push core was discarded to avoid contamination. The black reduced sediment core, 20 cm in length, was sliced into layers by every two centimetres with a pushcore equipment (sample ID: RS_1 ~ RS_10). Another sediment core was collected at the same site by a deep-sea light weighted monitorable and controllable long-coring system8, and the sample layers of 0~300 cm below the seafloor (cmbsf) was collected from the sediment core and sliced into 35-cm subsamples (sample ID: RS_11 ~ RS_19). All subsamples were stored at −80 °C until DNA extraction. Environmental data (CH4, H2S and SO42−) were detected in situ by a deep-sea laser Raman spectrometer mounted with the ROV in the previous report5,9.
DNA extraction
A schematic overview of workflow in this study was shown in Fig. 1b. The genomic DNA from 2.5 g of each sediment subsamples was extracted using the PowerSoil DNA Isolation Kit (QIAGEN). The genomic DNA from the 0.22μm filters was extracted using the PowerWater DNA Isolation Kit (QIAGEN). The DNA were examined by gel electrophoresis, and the concentration of DNA was measured using Qubit® dsDNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA). OD value is between 1.8~2.0, DNA contents above 0.4 μg are used to construct library (Table 2).
Metagenome sequencing
Metagenomic sequencing were performed at the Novogene (Tianjin, China) using the Illumina 2 × 150 PE protocols on an Illumina HiSeq X Ten platform. Preprocessing the Raw Data obtained from the sequencing platform using Readfq v8 (https://github.com/cjfields/readfq) was conducted to acquire the Clean Data for subsequent analysis. Clean Data of all 23 samples are available at NCBI Genbank (SRA) under the accession numbers SRR13892585~SRR13892607 (Table 2), and within the BioProject accession number PRJNA707313.
Genome binning
The initial de novo assembly was carried out using MEGAHIT v1.1.3 with default parameters10. Short genomic assemblies ( < 1,000 bp) that could have biased the subsequent analysis were first excluded. Genomes were then binned based on their tetranucleotide frequency, differential coverage, and GC content, as well as codon usage, using different binning tools, including MetaBAT 2, MaxBin 2.0 and CONCOCT implemented by MetaWRAP v1.2.1 pipeline (default parameters) (Supplementary Table 1)11,12,13. The binning results were refined using the MetaWRAP package (parameters: -c 60 -x 20)14 and all the produced bin sets were aggregated and dereplicated at 95% average nucleotide identity (ANI) using dRep v2.3.2 (parameters: -comp 60 -con 20 -sa 0.9)15. Taxonomic classification of each bin was determined by CheckM v1.0.3 and GTDB-Tk with default parameters (Supplementary Table 2)16,17. The bin quality assessment (completeness > 60% and contamination < 20%) of different binners was then performed by CheckM v1.0.3 (parameters: lineage_wf)17. Next, the selected bins for each sample were reassembled by using metaSPAdes implemented through the MetaWRAP pipeline14,18. The coding regions of the final MAGs were predicted with the the Prodigal v2.6.3 (metagenome mode -p meta)19. All the predicted genes were searched against the nr database and KEGG prokaryote database using diamond blastp (parameters: -e 1e-5–id 40)20,21. Data of all MAGs are available at NCBI Assembly under the accession numbers JAGLBO000000000~ JAGMFB000000000 (Supplementary Table 1).
Phylogenomic analysis
The 768 draft genomes and the 208 reference genome sequences accessed from NCBI GenBank (Supplementary Table 3) were combined to find orthologs for phylogenetic analysis by Orthofinder (default parameters)22. Each ortholog was aligned using MUSCLE v.3.8.31 (parameters:–maxiters 16)23, trimmed using trimAL v.1.2rev59 (parameters: -automated1)24 and manually assessed. Gene tree of each ortholog was constructed using FastTree v2.1.9 (parameters: -gamma -lg;)25. The final species tree was inferred based on 40,080 gene trees using STAG v1.0.0 (https://github.com/davidemms/STAG) and was viewed and annotated using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) (Fig. 2).
Data Records
This project has been deposited at DDBJ/ENA/GenBank under the BioProject accession no. PRJNA707313, with the Sequence Read Archive deposited under the accessions SRR13892585~SRR1389260726,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48. Other data is available through figshare49, including the fasta files containing the contigs of all 768 MAG, the newick format of the phylogenetic tree.
Technical Validation
Potential contamination of samples was limited by following guidelines for analyses of microbiota communities50,51. Briefly, the samples were pre-treated in a sterile station in the lab of the Research Vessel KEXUE. DNA extractions took place within a dedicated laboratory space under a laminar flow hood using aseptic techniques (such as, surface sterilisation, DNA-OFF, use of sterile plasticware, and use of aerosol barrier pipette tips). Sample processing was completed within 2 days, using the same batch of PowerSoil DNA Isolation Kit for all sediment samples, and PowerWater DNA Isolation Kit for all water-filters samples. The filtered and trimmed Illumina reads were evaluated for their sequencing qualities using fastp v0.20.1 (https://github.com/OpenGene/fastp) with default parameters52. In all samples, the Q score for the reads of each sample was calculated and showed that more than 90% of reads scored Q30 (Table 2), indicating that most of the reads were constructed with low error rates. Metagenome data have been assembled and refined into MAGs using the automated quality control steps and assembly procedures described in the manuscript. To ensure the assembly quality of the contigs, several kmers (21,29,39,59,79,99,119,141) were selected in the assembly procedures of MEGAHIT. As for binning, more strict standards were selected, and the sequence after binning was re-assembled to ensure the best result.
Code availability
The above methods indicate the programs used for analysis within the relevant sections. The code used to analyse individual data packages is deposited at https://github.com/zhcosa/MAGs-from-cold-seep.
References
Ceramicola, S., Dupré, S., Somoza, L. & Woodside, J. in Submarine Geomorphology (eds Aaron Micallef, Sebastian Krastel, & Alessandra Savini) 367-387 (Springer International Publishing, 2018).
Ruff, S. E. et al. Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. USA 112, 4015–4020 (2015).
Feng, D. et al. Cold seep systems in the South China Sea: An overview. J. Asian Earth Sci. 168, 3–16 (2018).
Zhang, X. et al. In situ Raman detection of gas hydrates exposed on the seafloor of the South China Sea. Geochem. Geophy. Geosy. 18, 3700–3713 (2017).
Zhang, X. et al. Development of a new deep-sea hybrid Raman insertion probe and its application to the geochemistry of hydrothermal vent and cold seep fluids. Deep-Sea Res. Pt. I 123, 1–12 (2017).
Cao, L. et al. In situ detection of the fine scale heterogeneity of active cold seep environment of the Formosa Ridge, the South China Sea. Journal of Marine Systems 218, 103530 (2021).
Du, Z., Zhang, X., Xue, B., Luan, Z. & Yan, J. The applications of the in situ laser spectroscopy to the deep-sea cold seep and hydrothermal vent system. Solid Earth Sciences 5, 153–168 (2020).
Wang, B. et al. A novel monitorable and controlable long-coring system with maximum operating depth 6000 m. Marine Sciences 42, 25–31 (2018).
Du, Z. et al. In situ Raman quantitative detection of the cold seep vents and fluids in the chemosynthetic communities in the South China Sea. Solid Earth Sciences 5, 153–168 (2018).
Li, D., Liu, C. M., Luo, R., Sadakane, K. & Lam, T. W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019).
Nissen, J. N. et al. Improved metagenome binning and assembly using deep variational autoencoders. Nat. Biotechnol. 39, 555–560 (2021).
Wu, Y. W., Simmons, B. A. & Singer, S. W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016).
Uritskiy, G. V., DiRuggiero, J. & Taylor, J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158 (2018).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017).
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892585 (2022).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892586 (2022).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892587 (2022).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892588 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892589 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892590 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892591 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892592 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892593 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892594 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892595 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892596 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892597 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892598 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892599 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892600 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892601 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892602 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892603 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892604 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892605 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892606 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR13892607 (2021).
Zhang, H. et al. Metagenome sequencing and 768 microbial genomes from cold seep in South China Sea, figshare, https://doi.org/10.6084/m9.figshare.16625644.v1 (2022).
Eisenhofer, R. et al. Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations. Trends Microbiol. 27, 105–117 (2019).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Acknowledgements
We acknowledge the support of the Research Vessel KEXUE of the National Major Science and Technology Infrastructure from the Chinese Academy of Sciences (CAS), and Canter for Ocean Mega-Science, CAS. We are especially grateful to the pilots and crew of FAXIAN ROV. We also thank all the laboratory members for their technical advice and helpful discussions. This work was funded supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2022QNLM030004-3), the National Natural Science Foundation of China (42030407 and 42076091) and the Senior User Project of RV KEXUE (KEXUE2021GH01 and KEXUE2019GZ06).
Author information
Authors and Affiliations
Contributions
M.W., H.Z. and C.L. designed the study. M.W., H.Z., H.C., L.C., C.L. and Z.Z. collected the samples. M.W., H.Z., H.C., H.W. and L.Z. performed the analysis. H.Z. and M.W. wrote the paper and prepared the figure and tables. All co-authors commented on the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, H., Wang, M., Wang, H. et al. Metagenome sequencing and 768 microbial genomes from cold seep in South China Sea. Sci Data 9, 480 (2022). https://doi.org/10.1038/s41597-022-01586-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-022-01586-x
- Springer Nature Limited
This article is cited by
-
Recovery of 1887 metagenome-assembled genomes from the South China Sea
Scientific Data (2024)
-
Trait biases in microbial reference genomes
Scientific Data (2023)
-
Mining of novel secondary metabolite biosynthetic gene clusters from acid mine drainage
Scientific Data (2022)