Abstract
Rheumatoid arthritis (RA) is a chronic inflammatory disorder with poorly defined aetiology characterised by synovial inflammation with variable disease severity and drug responsiveness. To investigate the peripheral blood immune cell landscape of early, drug naive RA, we performed comprehensive clinical and molecular profiling of 267 RA patients and 52 healthy vaccine recipients for up to 18 months to establish a high quality sample biobank including plasma, serum, peripheral blood cells, urine, genomic DNA, RNA from whole blood, lymphocyte and monocyte subsets. We have performed extensive multi-omic immune phenotyping, including genomic, metabolomic, proteomic, transcriptomic and autoantibody profiling. We anticipate that these detailed clinical and molecular data will serve as a fundamental resource offering insights into immune-mediated disease pathogenesis, progression and therapeutic response, ultimately contributing to the development and application of targeted therapies for RA.
Measurement(s) | Transcriptomics, small RNA, proteomics, autoantibodies, metabolomics, genotype, blood, urine |
Technology Type(s) | microarray, RNA sequencing, somascan, NMR metabolomics, luminex autoantibody array, exome array |
Sample Characteristic - Organism | Homo sapiens |
Similar content being viewed by others
Background & Summary
Rheumatoid arthritis (RA) is an immune-mediated inflammatory disease (IMID) that clinically manifests in the joints, but is systemic in impact. Early and intensive treatment is a critical determinant of long-term outcome, although clinical remission remains a minority outcome and sustained drug-free remission remains rare1.
The RA-MAP Consortium is a UK industry-academic precision medicine partnership funded by the Medical Research Council and the Association of the British Pharmaceutical Industry (ABPI). RA-MAP’s goals are to investigate clinical and biological predictors of disease outcome and treatment response in RA, using deep clinical and multi-omic phenotyping (Fig. 1). The study is in part motivated to inform the study design and analysis of future studies of blood and immune cell subsets in RA and other IMIDs. RA-MAP patients follow the UK-NHS standard of care, with first-line treatment with conventional synthetic disease-modifying anti-rheumatic drugs (csDMARD), such as methotrexate, which have slow onset of action. In the case of non-responders to csDMARDs, prolonged periods of uncontrolled disease activity can lead to joint damage and disability. Thus, RA-MAP seeks to address a major unmet need to identify patient-level predictors of response in order to identify patients with a greater or lesser chance of clinically responding to csDMARD treatment. Such information could guide treatment choices, possibly supporting fast-track biologic therapy, leading to improved long-term outcomes for patients, and saving time and money in achieving sustained disease control and improving the efficiency of clinical trials.
Molecular profiling of whole blood and peripheral blood mononuclear cells (PBMCs) has been widely used to investigate the molecular heterogeneity and pathogenesis of RA across a number of transcriptome analysis platforms2,3,4,5,6,7,8. However only a small number of studies have profiled more than 100 patients9,10,11. Most studies have focused on patients with well-established disease who were already on DMARD therapy. Although findings of interest have been identified in these studies, such as association between therapeutic response and type I interferon signatures8, there remains a major unmet need for better molecular characterization of both heterogeneity and disease activity in RA without the confounding effects of immunomodulatory therapy.
The “Towards A Cure for Early Rheumatoid Arthritis” (TACERA) study12 was designed to detect distinct disease subtypes and the immunological correlates of disease activity and autoimmunity in patients with early, seropositive RA, with a secondary objective to investigate biomarkers of initial drug response to methotrexate. The study had an additional exploratory objective, to compare the immune profiles and response seen in RA with innate immune profiles seen in healthy subjects prior to and subsequent to hepatitis B immunization. Although the RA and vaccine patients were not control matched, use of a common omics platform and bio-resource was initiated to enable a specific investigation of the innate immune response in RA, by comparison of the dysregulated immune response in early RA with a healthy innate immune response to the Hepatitis B antigen. To this end, we recruited a cohort of 267 early RA patients, and 52 healthy subjects receiving the hepatitis B vaccine. Following sample quality control exclusions we transcriptionally profiled 242 RA patients and 37 healthy vaccine recipients utilising whole blood, PBMCs, and CD14+, CD8+, and CD4+ leukocyte subsets at their first visit to the clinic and after 6 months, totalling 2257 unique samples. We ultimately followed these patients longitudinally for up to 18 months, collecting rich bio-samples and a range of clinical and omic data on the genome, transcriptome, proteome, metabolome and autoantibodies. The RA-MAP project has generated an unparalleled range of data and insights into the molecular heterogeneity of RA phenotypes in peripheral blood, which can serve as a fundamental reference for analysis of the blood immunological landscape in RA and other IMIDs.
Methods
Patient characteristics and study design
In the TACERA study, two hundred and seventy five patients were recruited of whom 270 fulfilled all eligibility criteria, that is newly diagnosed patients of at least 18 years of age with symptom duration less than 12 months, untreated with DMARDs or corticosteroids and who fulfilled either the 1987 American College of Rheumatology (ACR) or 2010 ACR/European League Against Rheumatism (EULAR) classification criteria for RA. Two eligible patients withdrew at baseline without providing any clinical information. A further patient who withdrew at baseline had some clinical information but insufficient to calculate disease activity scores. Therefore, for our cohort summary in Table 1 we describe baseline characteristics of these 267 remaining patients. Note that 239 of these 267 eligible patients had a 6-month assessment visit. All patients were seropositive at baseline: 93% were rheumatoid factor (RF) positive, and 87% were citrullinated protein antibody positive (ACPA). Subjects were recruited from 28 participating centres across the UK. Following enrolment subjects received treatment at the discretion of the supervising rheumatologist per the National Institute for Health and Clinical Excellence (NICE) guidelines for the management of RA in adults13. Patients were followed up for 18 months and seen every 3 months. Based on EULAR response criterion, 47.2% patients achieved a good response, 33.9% a moderate response and 18.9% showed no response. Clinical, laboratory, lifestyle, comorbidities and associated medication, patient reported outcome measures, and biological samples were collected at each visit. Blood samples for RNA extraction were taken at baseline and 6 months. Clinical data is summarised in Table 1. Ethical approval was authorised by the National Research Ethics Service London Central Committee (Reference number: 12/LO/0469). Informed, written consent was obtained from all study participants. Complete phenotypes and further details on the TACERA study are published elsewhere14,15.
Vaccine study design
Vaccine (Engerix B (recombinant Hepatitis B surface antigen)) recipients were recruited from healthcare workers receiving hepatitis B screening as part of their workplace induction at 4 participating centres across the UK (Newcastle, Birmingham, London and Glasgow). Subjects received Hepatitis B vaccination (20 micrograms Engerix B by IM injection at 0, 1 and 2 months), and were followed up for 8 visits (day -7, 0 (baseline), 1, 3, 7, 56, 57, 63). Clinical, lifestyle, demographics, and biological samples were collected at each visit. Blood samples for RNA extraction were taken at all 8 visits, with transcriptomics performed to the TACERA transcriptomic protocol on day -7, 0, 3, 56 and 63. Clinical data is summarised in Table 2. Informed, written consent was obtained from all study participants (study protocol is available on the RA-MAP figshare16).
Patient biosampling, extraction, and biobanking
Biosamples, including blood, serum, plasma and urine were obtained from patients every 3 months for 18 months in the TACERA and Vaccine studies. The SOPs required that blood was drawn and stored locally for up to 30 minutes before transport to the local processing hub. According to SOPs serum, plasma and urine samples were processed no longer than 60 minutes after collection. PBMC processing, followed by isolation of CD4, CD8 and CD14 subsets, took place within 60 minutes of blood draw. Isolated cells were then lysed and frozen in QIAzol lysis reagent and stored at −80 °C for later RNA extraction. Samples were stored prior to RNA extraction until the last participant, last visit. For the whole blood RNA the blood was drawn into a Tempus tube and then incubated for 3 hours at room temperature then frozen at −20 °C for 24 hours before long term storage at −80 °C. Like the cell subsets RNA the whole blood RNA was extracted from these samples at the end of the study. Blood for RNA extraction was collected in Tempus blood RNA tubes (Applied Biosystems) and mixed by inverting. Additional blood sampling for isolation of PBMCs was collected into EDTA Vacutainer collection tubes (Becton Dickinson) and separated using Leucosep separation tubes (Greiner). Cell subsets were isolated from the peripheral blood using magnetic cell sorting. The Miltenyi MACS system was used to positively isolate CD14-expressing monocytes, CD4-expressing T cells and CD8-expressing T cells from the isolated PBMCs by following the manufacturer’s protocol (Miltenyi Biotech). The purified cells were lysed in QIAzol Lysis Reagent. Cell subset micro RNA was extracted using miRNeasy minikits (Qiagen) following the manufacturer’s protocol. Whole blood RNA was extracted from Tempus Blood tubes using MagMAX RNA isolation kits (Ambion) and then was subjected to removal of globin mRNA using GLOBINclear human 20 reaction kits (Ambion) following the manufacturer’s protocol. Multi-omic analysis of samples was performed at baseline and 6 months. All samples (n = 34,540) were deposited with UK Biocentre (Milton Keynes, UK) and are available to researchers on request to the RA-MAP sample access panel (https://research.ncl.ac.uk/ra-map/). Detailed SOPs are available in the RA-MAP figshare16. Some of the logistical processing challenges faced by the study are described elsewhere12.
Microarray mRNA sample analysis
For microarray analysis, amplified RNA was hybridized to Illumina HumanHT-12 V4.0 expression beadchips and scanned on an Illumina Beadstation 500. Illumina’s GenomeStudio version 2011.1 with the Gene Expression Module v1.9.0 was used to generate signal intensity values. TACERA samples were randomized across the analysis plates, with samples from same RA subject (baseline and 6-month time points) assigned within same plate. Vaccine samples were run in separate batches, but also randomized across analysis plates. Non-normalised control and sample probe data was exported from GenomeStudio.
mRNA transcriptome data analysis
To perform QC and exploratory data analysis (EDA), all mRNA sample data was imported to the R Limma package17 as a combined matrix to enable direct comparison of cell subsets. Where the goal is to isolate effects within a particular cell type, single cell type processing may be more optimal. Background correction and quantile normalization were performed using the Limma neqc function18, based on methods described by Shi et al.19. All probes with detection p-values of at most 0.05 in at least 100 samples were removed. This ensured that all remaining probes were expressed in a minimum of 100 samples, or just under 10% of all profiles. Probes were aggregated to gene level using the Limma avereps function to further reduce dimensionality and increase reproducibility20, using a simple mean aggregator as implemented in the limma package. After pre-processing, filters and exclusions, 18,562 genes were present in the analysis ready expression matrix. Dimensionality reduction by Principal component analysis (PCA) was used to check for outliers and unsupervised clustering effects. 12 samples were flagged as outliers in the dataset, we removed these from subsequent analysis (See full EDA and outlier markdown documents on GitHub).
Small RNA sample and transcriptome data analysis
For Small RNA sequencing analysis, amplified small RNA was sequenced on an Illumina HtSeq. 2500 unit with single read flow cells to a depth of at least 10 million 50-bp reads per sample. TACERA samples were randomized across the analysis plates, with samples from same RA subject (baseline and 6-month time points) assigned within same plate. Adaptors were clipped off the reads using Trimmomatic21 (version 0.33) and then aligned to the GRCh38 genome using Bowtie-2 (version 2.3.0)22. Next, alignments to miRNA reference were counted with the htseq-count function from HTSeq (version 0.6.1p2)23 and the miRBase annotation release 22.1. Prior to normalization, transcripts in the resulting count table were filtered to a mean count per million (CPM) of at least 2, and normalised using the EdgeR CalcNormFactors function24.
Plasma protein analyte analysis
Plasma samples were selected from 100 baseline patients with higher baseline disease activity (DAS28 > 4) who divided equally at the 6 month visit into 50 patients in remission (DAS28 < 2.6) and 50 with active disease (DAS28 > 4). Plasma samples from 40 healthy (vaccine) recipient (VC) subjects, were analyzed concurrently with the RA patient samples. 1310 analytes were measured in the selected plasma samples for baseline (RA, vaccine) and 6-month (RA) visits at SomaLogic, LLC (Boulder, CO USA) using SOMAscan v3.2 platform. RA and vaccine recipient samples were randomized across the analysis plates, with samples from same RA subject (baseline and 6-month time points) assigned within same plate. 124 analytes were flagged by the vendor for failing QC standards, leaving 1186 analytes available for analysis. Relative fluorescence unit (RFU) data were sequentially normalized for hybridization controls (internal standards per sample) to remove inter-run hybridization artifacts, median signal across all samples to remove other potential assay biases (assumes same total protein concentration across sample set), and calibration controls (common sample standards across analysis plates). The normalized RFU values were log2-transformed and then each analyte was independently 0-centered to the mean of the healthy subject cohort by shifting. 2 samples failed the vendor’s QC standards for median normalization scale factors within range of 0.4 to 2.5 and were excluded from further analysis (both 6-month samples from the active disease group).
Auto-antibody sample analysis
501 serum samples were analysed from the TACERA cohort, comprising 265 baseline samples and 235 6-month follow-up samples. In parallel, 44 baseline and 38 follow-up samples from Vaccine recipients were measured. All samples were distributed on 96-well assay plates applying a randomised block design (timepoint, age, gender, healthy, RA).
A Luminex bead-based antigen array was produced (Protagen AG, Switzerland) to measure the autoantibody response against 192 human protein antigens. Antigens were selected based on literature data and autoantibody reactivity data of previous high-content profiling studies in RA and other rheumatic diseases. A subset of protein antigens (n = 46) were citrullinated in vitro using peptidyl arginine deiminase (PAD) to compare the autoantibody reactivity towards citrullinated and corresponding uncitrullinated antigens in early RA patients. Briefly, proteins were produced in Escherichia coli as His-tagged fusion proteins and purified by immobilised metal affinity chromatography. Coupling of antigens to magnetic carboxylated colour-coded beads (MagPlex microspheres, Luminex Corporation, Austin, Texas) was performed according to manufacturers’ protocols. Beads coupled with BSA, human IgG (hIgG), E. coli lysate and the eluate of vector only transformed E. coli were used as internal quality controls to evaluate the background reactivity, the measurement range or patient anti-E. coli reactivity, respectively. Finally, beads were combined and stored at 4–8 °C until use. An aliquot of the bead mix was incubated with the 1:100 diluted patient serum sample. Bound antibodies were measured following incubation with a secondary PE-labelled anti-human-IgG antibody in a FlexMap3D instrument (Luminex Corporation, Austin, Texas). The IgG reactivity values are given as median fluorescence intensity (MFI) and data of antigens fulfilling the minimum bead count criterion (>10 beads measured per bead ID) was used for data analysis. To monitor the inter-assay coefficient of variation, three in-process control samples were measured in triplicate on each 96-well assay plate using the autoantibody MFI values of all measured antigens. The overall median inter-plate CV was 7.7%. Evaluation of the control beads showed that the MFI values of control beads was as expected: The background reactivity towards BSA was 4 MFI, the reactivity to the E. coli lysate was 8 MFI, to the vector only eluate 6 MFI and to hIgG 22,000 MFI. The hIgG coupled bead was used to confirm the reactivity of the PE-conjugated detection antibody. To obtain reliable MFI values for data analysis the bead count statistics of the autoantibody data were evaluated. The median bead count of all samples was 167, with 0.01% of all samples having a bead count <10 and 0.9% of all samples having a bead count of <35. All samples and antigens met the bead-count criterion.
Auto-antibody data analysis
Raw Luminex autoantibody measurement values were processed and analysed using the R programming language (http://www.r-project.org/ version 3.3.0) and KNIME 3.2 (https://www.knime.org/) to produce text CSV files for quality control and further statistical analysis. The main pre-processing steps were removal of data points that did not pass the quality control with regard to bead count criterion, MFI values were transformed into log2 values. Following the exclusion of autoantibodies with <10% seropositivity in RA patients, 163 autoantibodies were retained for analysis. To adjust for systematic variation in the overall MFI values of individual samples, the data were median-centred by the sample.
Metabolomic sample preparation
After thawing, TACERA serum samples were centrifuged at high speed (13000 ×x g(av)) and then filtered by centrifuging at 10,000 × g(av) through a thoroughly pre-washed 3000 molecular weight cut-off filter (Pall, Omega 3k) to remove proteins which greatly improves the quality of the subsequent NMR spectra25. The filtrate was diluted 1:4 with D2O/H2O (40%) containing NaCl (150 mM), deuterated 4,4-dimethyl–4-silapentane-1-sulfonic acid (D6-DSS) (2 mM) as a chemical shift standard, difluorotrimethylsilanyl phosphonic acid (DFTMP) (0.4 mM) as a pH indicator26 as suggested by Chenomx Inc, sodium azide (0.4%) and sodium phosphate (100 mM) pH 7.0. The samples were then stored frozen at −80 °C until analysed, at which time they were thawed and a sample (35 µl) transferred to a 1.7 mm NMR tube which was then capped. Urine samples were thawed, centrifuged and diluted 1:4 with the NMR buffer detailed above. Samples were then carefully pH adjusted to 7.0 until stable27 and then frozen at −80 °C until analysed.
Urine and Serum metabolomics data analysis
One-dimensional 1H NMR spectra were acquired at 300 K using a NOESY pulse sequence including water suppression with pre-saturation on a Bruker DRX 600 MHz NMR spectrometer equipped with a TXI 1.7 mm cryoprobe. 2D homonuclear (1J-resolved) spectra were also acquired for each sample and heteronuclear (1H_1H_13H TOCSY and C HSQC) spectra were also recorded for selected samples to aid spectral assignment. Samples were processed and data calibrated with respect to the DSS signal. Spectra were read into Metabolab28, in Matlab (version 2016a, The Mathworks, Natick, MA), and spectra were segmented into 0.006-ppm (3 Hz) chemical shift ‘bins’ and the spectral area within each bin was integrated. Spectra were corrected for baseline offset using a spline fit and then normalised using Probabilistic Quotient Normalisation (PQN)29,30 and a generalised log transformation was applied31,32. Binned data were then compiled into a matrix, with each row representing an individual sample. NMR databases (Human Metabolome Database version 3) and the Chenomx NMR suite (Chenomx, Professional version 8.0)33 were used to identify and quantify metabolites present in each sample. Automated metabolite identification with Chenomx was used to produce an initial fit and then manual fitting of the Chenomx provided metabolite spectral library was done. This focussed on a set of metabolites previously identified as being present in human serum34 and urine35. Other published data on metabolites identified in human sera were also used to guide identification36,37. Following automated and manual metabolite identification, 40 known metabolites were identified and quantified in sera and 42 in urine.
Genotyping
Genotyping was performed using the Illumina HumanCoreExome-24-v1-0 (Batch 1) or Illumina InfiniumCoreExome-24-v1-1 (Batch 2) according to the manufacturer’s SOP. Raw intensity data files (idat format) from the Illumina iScan instrument were imported into GenomeStudio (v2011.1). Samples <90% call rate were excluded. Data was exported to PLINK PED/MAP format on the forward strand. Data was converted from PED/MAP to BED/BIM/FAM using PLINK v1.07. HumanCoreExome-24v1-0_A_PopulationReport_MAF_022015.txt or InfiniumCoreExome-24v1-1_A_PopulationReport_MAF.txt was used to obtain a list of all variants on the array with a MAF >0.005. This list was used to extract the variants from the genotype file. Variants with a GenomeStudio Cluster Separation <0.3 were excluded. Variants with <98% call rate were excluded. Individuals with <98% call rate were excluded. Palindromic SNPs (AT/CG) were excluded and the file split per chromosome. Variant IDs were updated to match Haplotype Reference Consortium v1.1 using GenotypeHarmonizer. SNPs in common between the two arrays were extracted from each dataset and the files combined (TACERA_combined). W.Rayner’s script HRC-1000G-check-bim-4.23.pl was used to align SNPs to HRC v1.1 panel. PLINK v1.9 was used to convert the PLINK files to VCF. The Wellcome Trust Sanger Institute imputation server was used to impute the data to the HRC v1.1 panel using SHAPEIT2 for phasing and PBWT for imputation.
HLA imputation methods
HLA imputation was performed using PLINK to extract 28Mb-34Mb on chromosome 6. SNP2HLA was used for imputation with the Type 1 Diabetes Genetics Consortium (T1DGC) Panel38. Custom bash and STATA scripts were used to extract the HLA haplotypes from the bgl.phased file.
Data Records
Figure 2 summarises the number of samples processed on each omic platform. All raw data and processed mRNA and small RNA data are available in the NCBI GEO series accession number GSE9747639. Serum and urine metabolomic data have been deposited at the MetaboLights database of the European Bioinformatics Institute (EBI) under MTBLS149740. Genotype data has been deposited in the European Genome-phenome Archive (EGA) with ID number EGAS0000100442441. De-Identified clinical data, X-ray, Somalogic proteomic data, including aptamer annotation and Protagen autoantibody data are available in the RA-MAP project in figshare16.
Technical Validation
Exploratory data analysis across omic platforms
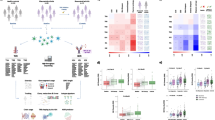
Whole blood, cell subsets and serum readouts from each individual omic data platform were compared at baseline and six months, using a unified exploratory data analysis (EDA) approach employing a range of multi-dimensional visualisation and dimensionality reduction methods implemented in the bioplotr package (https://github.com/dswatson/bioplotr). The technical validity of each platform was explored using visualisations of mean variance / dispersion plots, density plots, subject similarity and principal component analysis (PCA). Full EDA markdown documents and data files for each platform are available in the RA-MAP GitHub. In Fig. 3, PCA is used to gain an overview of each omics platform at baseline and 6-months in whole blood and cell subsets in the TACERA early RA cohort (a, c-h) and across a range of time points in the vaccine cohort (b, k-o). Similar separation by cell type is evident in both RA and vaccine cohorts. After QC, including limited outlier removal, all Omics platforms appear relatively homogeneous with no unexpected structure in PCA projections. Some evidence of separation by time is seen in the RA cohort in whole blood and cell subset mRNA, serum miRNA, serum autoantibodies and urine metabolomics. Clear time separation is less evident in the Vaccine cohort. Collectively the EDA across each platform provides consistent evidence of technical validation. In order to identify those clinical features driving the observed multi-omic changes, giving some biological validation to the data, we performed unsupervised PCA Driver analysis (Fig. 4) which shows the degree of association between Principal Components (PC1-5; % PC effect indicated) and clinical variables in each multi-omic platform. The driver plot heatmap indicates the –log q-value of the association with each clinical and technical variable. Significant drivers are indicated by outline with an FDR threshold of 5%. Unlike the relatively non-specific PCA projections, PC driver plots allow direct evaluation of influence of different clinical and technical variables on variation of expression in the samples. In the TACERA cohort, the larger PCs, representing the largest source of variance, are closely associated with time and DAS28 and related disease activity scores in whole blood and cell subset mRNA and whole blood miRNA; and rather less closely associated with the other platforms. Notably time and DAS28 correlate closely with the exception of the urine metabolome where changes in time, including the joint strongest association seen in the dataset, are less well correlated with disease activity. This leads us to conclude that changes over time in the urine metabolome driven by PC2 are unlikely to be disease related. We note that region hub shows some correlation with measures of disease activity in some Omic platforms. Samples from 28 patient recruiting centres were processed in 7 regional hubs14, we hypothesize that this may reflect a higher proportion of more severe RA cases seen in tertiary referral centres. In the Vaccine cohort, in contrast to the highly dysregulated immune system seen in RA patients, the disturbance following a vaccine related immune challenge in the healthy volunteers appears negligible.
Principal component analysis (PCA) across multiple omic platforms at baseline and 6-months in whole blood and cell subsets in (a) the TACERA early RA cohort and (b) across 6 visits (day −7, 0, 3, 56 and 63) in the vaccine cohort. Similar separation by cell type is evident in both RA and vaccine cohorts. Some evidence of separation by time is seen in the RA cohort, but this is less evident in the Vaccine cohort. Multi-omic PCA comparison at baseline and 6-months in early RA across (c) whole blood mRNA, (d) micro RNA (serum), (e) proteome (plasma), (f) autoantibodies (serum), (g) metabolome (serum) and (h) metabolome (urine). Clear separation with time is seen in Whole Blood and cell subset mRNA, Serum miRNA, Serum Autoantibodies and Urine metabolome.
Unsupervised PCA Driver analysis of multi-omic compartments across TACERA early RA patients and Hepatitis B vaccine recipients, showing clinical features and their degree of association with Principal Components (PC) 1–5, percentage indicating variation accounted for by the PC and with coloring indicating the –log q-value of the association (scaled to maximum range of PC1 across all compartments, off scale associations are indicated in grey with –log q written). Significant drivers with an FDR threshold of 5% are indicated by outline. Specific drivers of variation in expression in analysed samples are indicated in TACERA patients in (a) Whole Blood mRNA, (b) PBMC mRNA, (c) CD4 mRNA, (d) CD8 mRNA, (e) CD14 mRNA, (f) Whole Blood micro RNA, (g) Plasma proteomics, (h) Serum autoantibodies, (i) Serum metabolome, (j) Urine metabolome. In VACCINE recipients in (k) Whole Blood mRNA, (l) PBMC mRNA, (m) CD4 mRNA, (n) CD8 mRNA, (o) CD14 mRNA. Clinical features include XRAY (quantitative measure of bone erosion at sample timepoint (0 or 6 m)), TIME (sample annotation at baseline or 6-months), SYMP_DUR (Symptom duration at diagnosis), SMOKER (smoking status Y, N, Previous), SEX (M/F), RF (Rheumatoid factor positive Y/N), PAIN (quantitative measure of pain at sample timepoint (0 or 6 m)), FATIGUE (quantitative measure of fatigue at sample timepoint (0 or 6 m)), ETHNICITY (Ethnic origin), DAS28 (disease activity score in 28 joints at sample timepoint (0 or 6 m)), CRP (quantitative measure of C-reactive protein at sample timepoint (0 or 6 m)), BMI (Body Mass Index at baseline), ALCOHOL (Y/N), AGE (Age at baseline), ACPA (anti-citrullinated protein antibody positive Y/N), SEROLOGY (Hepatitis B serology at week 9). In the TACERA cohort in 3a-j the first two PCs are closely associated with DAS28 scores in Whole Blood, PBMC mRNA, CD4 mRNA, and CD14 mRNA; and rather less closely associated with the other platforms. In 3k-o, in contrast to the highly dysregulated immune system seen in RA patients, the biological perturbation following a vaccine-related immune challenge in the healthy volunteers appears negligible.
Collectively insights from EDA in the TACERA and Vaccine cohorts support the high technical quality of both data sets. Principal Component Driver analysis provides biological evidence of disease signatures in the TACERA cohort that are guiding our ongoing analysis of this dataset.
Usage Notes
Genotype data is made available through the European Genome-phenome Archive (EGA). Request for data access will be referred directly to our Data Access Committee: https://ega-archive.org/datasets/EGAD00001006736. If you need to request access to this data set, please contact the RA-MAP Data Access Committee (Contact person:m.r.barnes@qmul.ac.uk). Applicants will be asked to complete the Data Access Agreement (DAA) (including a brief summary of the proposal, proposed usage of the dataset, the storage of data, so the DAC can determine if the planned usage falls within the consents) and to agree to the terms and conditions of the DAA. The DAA must be signed by the applicant and the relevant Head of Department, or equivalent. If applications include a named collaborator then the collaborator’s Institution must sign and submit a separate DAA. A template DAA can be found on the EGA website: https://ega-archive.org/submission/dac/documentation.
Code availability
Fully annotated Executable R scripts and R Markdown documents are available in our public RA-MAP GitHub in order to allow complete reproduction of our analysis workflow (https://github.com/C4TB/RA-MAP). All analyses were conducted in R version 4.0.5.
References
Singh, J. A. et al. 2012 update of the 2008 American College of Rheumatology recommendations for the use of disease‐modifying antirheumatic drugs and biologic agents in the treatment of rheumatoid arthritis. Arthritis Care. Res. (Hoboken) 64, 625–639 (2012).
Batliwalla, F. et al. Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun. 6, 388–397 (2005).
Julià, A. et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. Plos One 4, e7556 (2009).
Lequerré, T. et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res. Ther. 8, R105 (2006).
Raterman, H. G. et al. The interferon type I signature towards prediction of non-response to rituximab in rheumatoid arthritis patients. Arthritis Res. Ther. 14, R95 (2012).
Tasaki, S. et al. Multi-omics monitoring of drug response in rheumatoid arthritis in pursuit of molecular remission. Nat. Commun. 9, 1–12 (2018).
Teixeira, V. H. et al. Transcriptome analysis describing new immunity and defense genes in peripheral blood mononuclear cells of rheumatoid arthritis patients. Plos One 4, e6803 (2009).
Van der Pouw Kraan, T. et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann. Rheum. Dis. 66, 1008–1014 (2007).
Lewis, M. J. et al. Molecular portraits of early rheumatoid arthritis identify clinical and treatment response phenotypes. Cell Rep. 28, 2455–2470. e2455 (2019).
Nakamura, S. et al. Identification of baseline gene expression signatures predicting therapeutic responses to three biologic agents in rheumatoid arthritis: a retrospective observational study. Arthritis Res. Ther. 18, 159 (2016).
Walsh, A. M. et al. Integrative genomic deconvolution of rheumatoid arthritis GWAS loci into gene and cell type associations. Genome Biol. 17, 79 (2016).
Cope, A. P. et al. The RA-MAP Consortium: a working model for academia–industry collaboration. Nat. Rev. Rheumatol. 14, 53 (2018).
Deighton, C., O’Mahony, R., Tosh, J., Turner, C. & Rudolf, M. Management of rheumatoid arthritis: summary of NICE guidance. BMJ 338 (2009).
Tom, B. & Consortium, RA-MAP. Characterization of disease course and remission in early seropositive rheumatoid arthritis. medRxiv (2020).
Consortium, RA-MAP. Characterization of disease course and remission in early seropositive rheumatoid arthritis: Results from the TACERA longitudinal cohort study. Ther. Adv. Musculoskelet. Dis. 13, 1759720x211043977 (2021).
Consortium, RA-MAP. RA-MAP, a multi-omic immune cell landscape in early RA. Figshare https://doi.org/10.6084/m9.figshare.c.5491611.v1 (2021).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Smyth, G. K., Michaud, J. & Scott, H. S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075 (2005).
Shi, W., Oshlack, A. & Smyth, G. K. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 38, e204 (2010).
Miller, J. A. et al. Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinform. 12, 1–13 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Tiziani, S. et al. Optimized metabolite extraction from blood serum for H-1 nuclear magnetic resonance spectroscopy. Anal. Biochem. 377, 16–23 (2008).
Reily, M. D. et al. DFTMP, an NMR Reagent for Assessing the Near-Neutral pH of Biological Samples. J. Am. Chem. Soc. 128, 12360–12361 (2006).
Viant, M. R., Ludwig, C., Rhodes, S., Guenther, U. L. & Allaway, D. Validation of a urine metabolome fingerprint in dog for phenotypic classification. Metabolomics 3, 453–463 (2007).
Ludwig, C. & Gunther, U. MetaboLab - advanced NMR data processing and analysis for metabolomics. BMC Bioinform. 12, 366 (2011).
Dieterle, F., Ross, A., Schlotterbeck, G. & Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290 (2006).
Kohl, S. M. et al. State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolomics 8, 146–160 (2012).
Viant, M. R. Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochem. Biophy. Res. Commun. 310, 943–948 (2003).
Parsons, H. M., Ludwig, C., Gunther, U. L. & Viant, M. R. Improved classification accuracy in 1- and 2-dimensional NMR metabolomics data using the variance stabilising generalised logarithm transformation. BMC Bioinform. 8, 234 (2007).
Weljie, A. M., Newton, J., Mercier, P., Carlson, E. & Slupsky, C. M. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 78, 4430–4442 (2006).
Psychogios, N. et al. The Human Serum Metabolome. Plos One 6 (2011).
Bouatra, S. et al. The human urine metabolome. Plos One 8, e73076 (2013).
Nicholson, J. K., Foxall, P. J. D., Spraul, M., Farrant, R. D. & Lindon, J. C. 750-MHz H-1 and H-1-C-13 NMR-Spectroscopy of Human Blood-Plasma. Anal. Chem. 67, 793–811 (1995).
Beckonert, O. et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2, 2692–2703 (2007).
Brown, W. et al. Overview of the MHC fine mapping data. Diabetes Obes. Metab. 11, 2–7 (2009).
NCBI Gene Expression Omnibus https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97476 (2021).
Young, S. P., Barnes, M. R., Lendrem, D., Fisher, B. & Isaacs, J. Metabolights https://www.ebi.ac.uk/metabolights/MTBLS1497 (2021).
European Genome Phenome Archive https://ega-archive.org/datasets/EGAD00001006736 (2020).
Acknowledgements
The programme of research described in this article is funded by the Medical Research Council, UK (An Immunological Toolkit for Clinical Application, grant number G1001518 and Towards a cure for early rheumatoid arthritis, grant number G1001516). The RA-MAP Consortium would particularly like to thank members of the MRC Immunity and Inflammation Stratified Medicine Steering Group and Officers of the Medical Research Council who have supported the work of RA-MAP from the outset. In addition, the RA-MAP team acknowledges the exceptional contributions and talents of Christopher John (1986–2020), Meilien Ho (1960–2016) and Sally Hollis (1967–2019). This project was enabled through access to the MRC eMedLab Medical Bioinformatics infrastructure supported by the Medical Research Council [grant number MR/L016311/1]. SV is supported by Versus Arthritis (grant numbers 20385, 20380) and the NIHR Manchester Biomedical Research Centre, UK. JDI is a National Institute for Health Research (NIHR) Senior Investigator and his work is supported by the Research into Inflammatory Arthritis Centre Versus Arthritis and the NIHR Newcastle Biomedical Research Centre in Ageing and Long-term Conditions. The views expressed are those of the author(s) and not necessarily those of the National Institute for Health Research NIHR or the Department of Health and Social Care.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptualisation, M.R.B., C.P., C.J., M.E., A.C., D.L. and J.I.; Formal Analysis, C.J., D.W., S.R., G.S., M.J.L., P.B. J.C., S.N., R.H., J.T., D.L., B.T.; Investigation; Data Curation, C.J. and M.R.B.; Writing Original Draft, M.R.B., C.J., M.E. and S.B.; Writing Review and Editing, C.J., M.E., M.R.B., C.P., A.C., J.I., B.F., F.P., R.L., C.C., D.L., G.S., G.A. and M.D.; Project Administration, S.B. and A.P.; Funding Acquisition, A.C., C.P., I.M., C.B., M.E. and J.I. All authors read and commented on drafts of the manuscript and approved the final submitted manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.R.B. declares that he has acted as a consultant for or received honoraria or funding from Janssen, GSK, Servier, United Healthcare Group and Eli Lilly. J.D.I. declares that he has acted as a consultant for or received honoraria from Pfizer, Lilly, Abbvie, Janssen, Celltrion, Epirus, Boehringer, Baxalta. For a full list of competing interests for all RA-MAP co-authors, see Supplementary Table 1.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
The RA-MAP Consortium. RA-MAP, molecular immunological landscapes in early rheumatoid arthritis and healthy vaccine recipients. Sci Data 9, 196 (2022). https://doi.org/10.1038/s41597-022-01264-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-022-01264-y
- Springer Nature Limited
This article is cited by
-
FOXP3 splice variant expression in males and females in healthy populations and in kidney transplant recipients
Scientific Reports (2024)
-
A unified metric of human immune health
Nature Medicine (2024)
-
Precision medicine: the precision gap in rheumatic disease
Nature Reviews Rheumatology (2022)