Abstract
A few studies indicate that nitrate can reduce dysbiosis from a periodontitis point of view. However, these experiments were performed on samples from healthy individuals, and it is unknown if nitrate will be effective in periodontal patients, where the presence of nitrate-reducing bacteria is clearly reduced. The aim of this study was to test the effect of nitrate and a nitrate-reducing R. aeria (Ra9) on subgingival biofilms of patients with periodontitis. For this, subgingival plaque was incubated with 5 mM nitrate for 7 h (n = 20) or 50 mM nitrate for 12 h (n = 10), achieving a ~50% of nitrate reduction in each case. Additionally, Ra9 was combined with 5 mM nitrate (n = 11), increasing the nitrate reduced and nitrite produced (both p < 0.05). The addition of nitrate to periodontitis communities decreased biofilm mass (50 mM > 5 mM, both p < 0.05). Five millimolar nitrate, 50 mM nitrate and 5 mM nitrate + Ra9 led to 3, 28 and 20 significant changes in species abundance, respectively, which were mostly decreases in periodontitis-associated species. These changes led to a respective 15%, 63% (both p < 0.05) and 6% (not significant) decrease in the dysbiosis index. Using a 10-species biofilm model, decreases in periodontitis-associated species in the presence of nitrate were confirmed by qPCR (all p < 0.05). In conclusion, nitrate metabolism can reduce dysbiosis and biofilm growth of periodontitis communities. Five millimolar nitrate (which can be found in saliva after vegetable intake) was sufficient, while increasing this concentration to 50 mM (which could be achieved by topical applications such as a periodontal gel) increased the positive effects. Ra9 increased the nitrate metabolism of periodontitis communities and should be tested in vivo.
Similar content being viewed by others
Introduction
In susceptible individuals, periodontal diseases can develop as a result of an inflammatory response to dental plaque accumulation1. As the biofilm becomes thicker and inflammation increases, environmental changes can stimulate dysbiosis by selecting for anaerobic, inflammation-tolerant, proteolytic and/or alkaliphilic species2. The initial stage of periodontal disease is gingivitis, which is a mostly reversible inflammation of the gums present in most adolescents3. Long-lasting or repeated episodes of gingivitis can lead to periodontitis, which is a chronic and destructive inflammatory disease. Periodontitis may affect up to 50% of the adult population, with ~10% suffering from severe periodontitis4.
Within the oral cavity, periodontitis can be painful in some cases, cause halitosis and ultimately lead to tooth loss. In addition, emerging evidence suggests that periodontitis’ consequences extend beyond the oral cavity. For example, periodontitis is associated with an increased risk of diabetes5, rheumatoid arthritis6, atherosclerosis7, and hypertension8. Current periodontitis treatments are resource-intensive, time-consuming, and often only partially successful9. Inflammation and dysbiosis are reduced after periodontal treatment, but frequently reappear over the following months. Thus, new strategies to improve treatment efficiency are required to reduce the global health and financial burdens of periodontitis. In this respect, novel treatments such as prebiotics and probiotics that reduce dysbiosis (or stimulate eubiosis) and decrease inflammation should be explored2,9.
Periodontitis-associated microbiota composition is diverse and includes different species of the phyla Bacteroidota (formerly known as Bacteroidetes), Candidatus Saccharibacteria (formerly Candidate Division TM7), Bacillota (formerly Firmicutes), Pseudomonadota (formerly Proteobacteria), Spirochaetes, and Synergistetes10. Some of the species with the strongest association with periodontitis include the periodontal “red complex” (Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia) in which different potential virulence mechanisms have been identified11. In 2014, Pérez-Chaparro et al.10 systematically reviewed the literature, identifying 17 species associated with periodontitis. In addition to an increase in disease-associated species, health-associated species decrease in periodontitis, and their proportion is recovered after periodontal treatment in most patients9. Recently, Chen et al. developed the subgingival microbial dysbiosis index (SMDI) (i.e., a machine learning-based index assessing periodontal health in the subgingival plaque bacteriome) that calculates the amount of dysbiosis based on the abundance of different disease- and health-associated species12. Interestingly, these authors found that nitrate has dysbiosis-lowering properties when applying their index to in vitro data from Rosier et al.13 and a clinical study by Jockel-Schneider et al.14. In the latter clinical study, nitrate intake by patients with chronic gingivitis also reduced gingival inflammation15.
Several potential mechanisms of nitrate have been described that could improve periodontal health (reviewed by Rosier et al.16). In short, oral bacteria reduce nitrate to nitrite and nitric oxide—a host signalling molecule with antimicrobial activity. On the one hand, nitric oxide could directly reduce inflammatory pathways of host cells. In addition, nitric oxide can kill periodontitis-associated species (e.g., Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans have been shown to be sensitive to nitric oxide)17. In contrast, periodontal health-associated Rothia and Neisseria have been found to correlate negatively with inflammatory cytokines18,19,20 and their increase in the presence of nitrate could reduce inflammation. Importantly, current studies focusing on nitrate and oral health have only included patients without periodontitis. It is therefore important to determine how nitrate affects the bacterial composition in periodontitis. Additionally, as it is known that nitrate-reducing bacteria (e.g., Rothia dentocariosa and Rothia aeria) decrease in periodontitis and can increase after periodontal treatment9,12,21, it should be tested if nitrate-reducing probiotics have additional benefits.
The aim of this study was therefore to test the effect of nitrate and a nitrate-reducing Rothia aeria CECT9999 (Ra9) strain on periodontitis communities in vitro. For this, fresh subgingival plaque samples of periodontitis patients were grown ex vivo in an impedance based system that measures real-time biofilm growth22. Subgingival biofilm growth was monitored under a physiologically relevant nitrate concentration of 5 mM in the presence or absence of the Ra9 strain, or a high nitrate concentration of 50 mM, which could be obtained by the topical application of nitrate. Supernatant samples were taken for the measurements of nitrate, nitrite and pH. The remaining biofilms were collected for DNA isolation and Illumina sequencing of the 16S rRNA gene. Finally, the effect of nitrate on periodontitis-associated species was further studied by monitoring changes in a 10-species biofilm model using qPCR.
Results
Effect of nitrate with or without probiotic candidate on periodontal biofilm growth
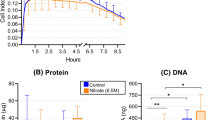
Subgingival plaque samples of periodontitis patients (patient information can be found in Supplementary Table 1) were incubated with nitrate. The incubation time for experiments with 5 mM (n = 20) and 50 mM nitrate (n = 10) was 7 h and 12 h (Table 1, Fig. 1), respectively. These timepoints were selected to reach a similar percentage (~50%) of nitrate reduction for each concentration of nitrate (Fig. 2A, B). Biofilm attachment and growth of in vitro periodontal plaque samples were monitored in real-time by impedance measurements under different growth conditions (Table 1). Figure 1 shows the average growth curve of the biofilms over time, expressed as Cell Index (CI) values22. Biofilm growth of communities grown in the presence of 50 mM nitrate was significantly lower than in the paired control samples at all timepoints after zero (all p < 0.05). In the case of the biofilms grown with 5 mM nitrate, a significant decrease in biofilm growth was observed from 1 h until the last timepoint (7 h, p < 0.05), indicating that the quantity of the final biofilms was lower.
Real-time 7 or 12 h growth curves of in vitro periodontal plaque biofilms under different treatment conditions, namely (A) samples supplemented with or without 5 mM nitrate (n = 20), or (B) with or without 50 mM nitrate (n = 10). Biofilm formation is expressed as Cell Index (CI) as determined using impedance measurements. Standard deviations are shown at 30 min intervals. The error bars represent the standard deviation of the mean. Statistically significant differences (p-value < 0.05 according to Wilcoxon matched pairs signed-rank test) are marked by an asterisk (*). The addition of the Ra9 strain appeared to affect initial biofilm formation, but had no significant effect on the final biofilm quantity (Supplementary Fig. 1).
Concentration of nitrate and nitrite in the supernatant of (A) biofilms supplemented with or without 5 mM nitrate (n = 20), after 7 h of biofilm growth, (B) with or without 50 mM nitrate (n = 10), after 12 h of biofilm growth, or (C) biofilms supplemented with or without 5 mM nitrate or 5 mM nitrate plus Ra9 (n = 11), after 7 h of biofilm growth. Supernatant pH of (D) biofilms supplemented with or without 5 mM nitrate, after 7 h of biofilm growth (n = 20), and (E) with or without 50 mM nitrate (n = 10), after 12 h of biofilm growth, or (F) biofilms (n = 11) supplemented with or without 5 mM nitrate or 5 mM nitrate plus Ra9, after 7 h of biofilm growth. In Fig. 2A–C the striped bar on the left side of the graph illustrates the concentration of nitrate added at baseline. Circles represent individual samples. The error bars represent the standard deviation of the mean. An asterisk (*) or distinct letters are used to indicate a statistically significant difference between treatment conditions in supernatant nitrite and pH (uppercase letters) or supernatant nitrate (Greek letters) (p-value < 0.05 according to Wilcoxon matched-pairs signed rank test, with Bonferroni correction for multiple comparisons for Fig. 2C, F). Ra9: Rothia aeria CECT9999.
Apart from the 5 mM and 50 mM nitrate conditions, the effect of a nitrate-reducing probiotic in the presence or absence of 5 mM nitrate was determined (n = 11). The addition of the probiotic candidate Ra9 with or without nitrate appeared to affect initial biofilm formation, but did not significantly affect the final biofilm quantity compared to the untreated control (Supplementary Fig. 1). When comparing the probiotic condition with and without 5 mM nitrate, no significant difference was observed in biofilm growth.
Effect of nitrate and probiotic candidate addition on denitrification and pH
To determine the nitrate reduction capacity (NRC) of the in vitro communities, nitrate and nitrite concentrations were determined in the supernatant of all samples collected after in vitro biofilm growth. Figure 2A–C show that nitrate, when supplied to the medium at a concentration of 5 mM (310 mg/L) or 50 mM (3100 mg/L), was partly reduced at the end of the biofilm growth period (7 h and 12 h respectively). These timepoints were selected to reach a similar percentage of nitrate reduction, leading to 56% reduction of the 5 mM nitrate (Fig. 2A) and 42% of the 50 mM nitrate (Fig. 2B) after 7 h and 12 h, respectively. The addition of the nitrate-reducing probiotic candidate significantly enhanced the nitrate reduction capacity of the periodontitis communities (Fig. 2C). Biofilms supplemented with 5 mM nitrate plus the probiotic reduced 97% of nitrate added after 7 h (p < 0.001 compared to the biofilms supplemented with 5 mM nitrate alone).
The reduction of nitrate to nitrite occurs at an equimolar ratio, i.e., a production of 5 mM (220 mg/L) nitrite from 5 mM (310 mg/L) nitrate, after which nitrite can be further reduced to form, for example, nitric oxide or ammonium16. Therefore, in parallel to the reduction in nitrate, an increase in the concentration of nitrite was observed in the 5 mM and 50 mM nitrate treatment conditions (both p < 0.05). After 7 h, 34% (i.e., ~1.7 mM) of nitrite produced had been further reduced in samples treated with 5 mM nitrate (Fig. 2A). In samples treated with 50 mM nitrate after 12 h, 10% (i.e., ~5 mM) of the nitrite produced had been further reduced (Fig. 2B). The increased nitrate reduction in the presence of the probiotic also increased the concentration of nitrite detected after 7 h compared to samples treated with 5 mM nitrate only (Fig. 2C).
A concentration of 5 mM nitrate caused a small but significant increase in pH (pH 6.6 ± 0.19 compared with 6.5 ± 0.33 of the control condition, p < 0.05), while no significant effect of 50 mM nitrate was observed on supernatant pH at the end of the biofilm growth period (Fig. 2D, E). The addition of the Ra9 strain in combination with 5 mM nitrate or the Ra9 strain alone resulted in a small but significant decrease in pH (pH 6.5 ± 0.09 and 6.3 ± 0.18 respectively, compared with pH 6.7 ± 0.10 of the control condition, both p < 0.01) (Fig. 2F).
The effect of nitrate and probiotic candidate addition on bacterial composition
The effect of nitrate and the Ra9 strain on the bacterial composition of the in vitro periodontal plaque biofilms was assessed by 16S rRNA gene sequencing analysis. Both in the control as well as in the 5 mM nitrate-treated biofilms a Streptococcus sp., Fusobacterium nucleatum, Porphyromonas gingivalis and a Veillonella sp. were the most dominant taxa (Fig. 3A). Similar species were found at high abundance in samples treated with 50 mM nitrate (a Streptococcus sp., Fusobacterium nucleatum, a Veillonella sp., and Veillonella parvula) (Fig. 4A). The bacterial composition of the periodontal biofilm samples reflects the clinical origin, as known periodontitis-related bacteria such as Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia (“red complex” bacteria) and/or other species of these genera were also well represented in the biofilms.
Bacterial composition of periodontal plaque biofilms grown in vitro supplemented with or without 5 mM nitrate (n = 20). A Relative abundance at the species taxonomic level as determined by 16 S rRNA gene sequencing. The top 20 most abundant species are shown and sorted by overall mean abundance; all other species are included as “other”. B Log2 fold-change of relative abundance of all species with statistically significant changes (ANCOM-BC adjusted p-value < 0.05). Species identified by the Boruta method as a significantly relevant feature are marked (#). C Box and whisker plot of the dysbiosis index. The dysbiosis index is expressed as SMDI score12. Plotted are the median, 25th to 75th percentile (box) and lowest to highest value (whiskers). A statistically significant difference (p-value < 0.05 according to Wilcoxon matched-pairs signed rank test) is marked by an asterisk (*).
Bacterial composition of periodontal plaque biofilms grown in vitro supplemented with or without 50 mM nitrate (n = 10). A Relative abundance at the species taxonomic level as determined by 16 S rRNA gene sequencing. The top 20 most abundant species are shown and sorted by overall mean abundance; all other species are included as “other”. B Log2 fold-change of relative abundance of all species with significant changes (ANCOM-BC adjusted p-value < 0.05). Species identified by the Boruta method as a significantly relevant feature are marked (#). C Box and whisker plot of the dysbiosis index. The dysbiosis index is expressed as SMDI score12. Plotted are the median, 25th to 75th percentile (box) and lowest to highest value (whiskers). A statistically significant difference (p-value < 0.05 according to Wilcoxon matched-pairs signed rank test) is marked by an asterisk (*).
Comparison of the general microbial composition (Supplementary Fig. 2) of biofilms receiving different treatments indicates no significant effect of the addition of nitrate or nitrate plus the probiotic candidate Ra9 on biofilm richness and evenness as expressed by Shannon diversity indexes (Supplementary Fig. 2D–F). Furthermore, CCA analysis showed no significant separation between control and nitrate treatment conditions (Supplementary Fig. 2A, B). For samples treated with Ra9 with or without nitrate, a significant separation between microbial communities is observed in the CCA plot (Supplementary Fig. 2C).
At the level of individual species, significant changes (all p < 0.05) can be observed between treatment conditions, shown as Log2 fold-changes in Figs. 3B, 4B and 5B. In general, disease-associated species appear to decrease when adding nitrate (50 mM > 5 mM) or nitrate combined with the Ra9 strain. Treatment using 5 mM nitrate (Fig. 3B) resulted in a decreased abundance of Porphyromonas gingivalis, Treponema maltophilum, and an unassigned Streptococcus sp., compared to controls. The 10-fold higher 50 mM nitrate treatment (Fig. 4B) increased the relative abundance of a Rothia sp. as well as an Aggregatibacter sp. (other than the known disease-associated species Aggregatibacter actinomycetemcomitans, which is detected separately and does not change significantly) and a Neisseria sp., whereas 25 species decreased in abundance, including multiple species of Fusobacterium, Porphyromonas, and Prevotella, among others (all p < 0.05). Specifically, in samples treated with 50 mM nitrate, the classified species that decreased significantly were Capnocytophaga leadbetteri, Fusobacterium nucleatum, Prevotella oris, Leptotrichia wadei, Granulicatella elegans, Dialister invisus, Tannerella forsythia, Alloprevotella tannerae, Eikenella corrodens, Filifactor alocis, Porphyromonas endodontalis, Prevotella intermedia and Porphyromonas pasteri. The addition of the probiotic candidate Ra9 in samples treated with 5 mM nitrate (Fig. 5B) also resulted in a significant increase of a Rothia sp. (though not classified, this is likely to correspond to Rothia aeria, resulting from the added Ra9 strain). Rothia mucilaginosa, a species that increased in samples treated with 50 mM nitrate, decreased in samples treated with both 5 mM nitrate and the probiotic (p < 0.05), possibly resulting from nutrient and niche competition with Rothia aeria. In addition, the abundance of 17 species decreased (all p < 0.05), among which we find disease-associated species such as Selenomonas sputigena, Slackia exigua and Fusobacterium nucleatum.
Bacterial composition of periodontal plaque biofilms grown in vitro supplemented with or without 5 mM nitrate and Ra9 (n = 11). A Relative abundance at species taxonomic level as determined by 16 S rRNA gene sequencing. The top 20 most abundant species are shown and sorted by overall mean abundance; all other species are included as “other”. B Log2 fold-change of relative abundance of all species showing significant changes (ANCOM-BC adjusted p-value < 0.05). Species identified by the Boruta method as a significantly relevant feature are marked (#). C Box and whisker plot of the dysbiosis index. The dysbiosis index is expressed as SMDI score12. Plotted are the median, 25th to 75th percentile (box) and lowest to highest value (whiskers). Differences are not statistically significant (adjusted p-value > 0.05 according to Wilcoxon matched-pairs signed rank test). Ra9: Rothia aeria CECT9999.
The effect of nitrate and probiotic candidate addition on the dysbiosis index
The dysbiosis level of the in vitro periodontal plaque biofilms was determined by computing the SMDI score12. Treatment of in vitro periodontal plaque biofilm with 5 mM nitrate or 50 mM nitrate significantly reduced the SMDI score compared to controls (Figs. 3C and 4C). This reduction was a 0.25 reduction (15%) in SMDI score for 5 mM nitrate (after 7 h) and a 0.35 reduction (63%) in SMDI score for 50 mM nitrate (after 12 h). Interestingly, when adding Ra9 in combination with 5 mM nitrate (synbiotic treatment), the SMDI only decreased 6% and this decrease was not significant, possibly resulting from Ra9 decreasing the abundance of health-associated species with a similar metabolism (e.g., Rothia mucilaginosa, which is one of the top health-associated species that affect the SMDI score12).
The effect of nitrate on a 10-species biofilm model
The sequencing results of periodontal plaque grown with nitrate indicated that nitrate could decrease the quantity of anaerobic bacteria associated with periodontitis. To confirm this possibility, a multi-species in vitro biofilm model containing a synthetic community of 10 oral strains was used to study the effect of nitrate on the growth of specific bacterial species. qPCR quantification of this 10-species community suggests that nitrate significantly reduced the growth of specific periodontitis-associated bacteria, including Fusobacterium nucleatum, Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis (all p < 0.05, Fig. 6).
qPCR quantification of total colony forming equivalents (CFE) of bacterial species from a 10-species in vitro periodontal biofilm model23 treated with 6.5 mM nitrate compared to control biofilms. In this model, Veillonella dispar and Actinomyces naeslundii are nitrate-reducing species. Significant differences (adjusted p-value < 0.05 according to Mann–Whitney U-test with Bonferroni–Dunn multiple testing correction) are marked by an asterisk (*). The error bars represent the standard deviation of the mean.
Discussion
Evidence from the last decade indicates that nitrate metabolism can stimulate eubiosis in individuals without periodontitis16. However, this is the first study in which the effect of nitrate on dysbiotic communities of patients with periodontitis was explored. Both 5 mM nitrate (a physiologically relevant concentration) and 50 mM nitrate (a concentration that could be obtained by the topical application of nitrate) decreased the biofilm quantity and dysbiosis index of the biofilms, indicating a beneficial effect on both bacterial growth and composition. A lower amount of subgingival plaque with a more health-associated composition is likely to reduce inflammation in vivo. The 50 mM concentration of nitrate led to more significant decreases in periodontitis-associated species and an additional reduction of biofilm accumulation compared with 5 mM nitrate. We suggest that the effect of nitrate-rich vegetable intake, as well as the topical application of nitrate in periodontal pockets that leads to a local high nitrate concentration (e.g., by using a periodontal gel with nitrate and antioxidants), should thus be explored in vivo. When adding the Rothia aeria CECT9999 (Ra9) strain to the 5 mM nitrate condition, more nitrate was reduced, and more nitrite was produced, which was accompanied by more significant changes in biofilm composition. An increase in nitrite production could improve systemic health by increasing systemic nitric oxide levels, which can improve conditions such as hypertension and diabetes—both associated with periodontitis5,8. This in vitro study indicates that nitrate as a prebiotic and nitrate together with a nitrate-reducing probiotic (i.e., a synbiotic combination) are promising (adjunct) treatments for periodontitis.
In this study, subgingival plaque samples of 30 periodontitis patients were divided in three experiments: 5 mM nitrate (n = 9), 5 mM nitrate in combination with the Ra9 strain (n = 11) and 50 mM nitrate (n = 10). All experiments contained relevant control conditions, and in the second group of 11 patients, a 5 mM nitrate condition without the Rothia aeria strain was added, leading to a total of 20 individuals treated with only 5 mM nitrate (Table 1). Subgingival plaque incubated with 5 mM nitrate was grown for 7 h (the same time as used previously for healthy communities by Rosier et al.24) and communities incubated with 50 mM nitrate were grown 12 h, both growth periods allowing for around 50% reduction of the different nitrate concentrations.
The final biofilm mass of the communities grown for 7 h with 5 mM nitrate was significantly reduced by 15% (p < 0.05), while after 7 h of growth, the biofilms for the communities grown with 50 mM were reduced by 53% (quantified in real time) (p < 0.05). After 12 h, the final cell index of the biofilms grown with 50 mM nitrate remained 42% lower than the control condition (p < 0.05). Dental plaque accumulation is considered the main stress factor in periodontitis development1,2,25. The host responds to the accumulated biofilms with gingival inflammation that can change the environment and give a selective advantage to periodontitis-associated proteolytic species1,2. This study indicates that nitrate could decrease the accumulation of dysbiotic periodontal plaque, which was not observed previously when adding nitrate to healthy communities13. Nitrate could reduce biofilm growth of sensitive bacteria because of the antimicrobial properties of its reduction products, such as nitric oxide17, which is also a biofilm dispersal signal for different bacteria2. Oral bacteria have been shown to have different tolerance levels of nitrate, nitrite and nitric oxide17,26, potentially explaining variations in responses of healthy biofilms compared with dysbiotic communities. Thus far this has been the only study testing the effect of nitrate on periodontal community growth. To our knowledge, there is only one study where the effect of nitrate intake on plaque accumulation was tested in vivo: Jockel-Schneider et al. tested the effect of nitrate-rich lettuce juice intake (compared to a placebo) on different clinical parameters and observed a trend of decrease in plaque control record15. The current data thus suggest that the effect of nitrate on plaque accumulation in patients with periodontal diseases should be further explored.
Apart from a decrease in biofilm quantity, the microbial composition of the biofilm affects inflammation, as some species stimulate inflammatory responses whereas others reduce inflammation27. In our study, both 5 mM nitrate and 50 mM nitrate significantly reduced the dysbiosis index in the final biofilms by 15% and 63%, respectively. This is in accordance with a previous study that showed that the SMDI dysbiosis index of healthy communities decreased after 5 h of growth with 6.5 mM nitrate in vitro12,13. Additionally, the dysbiosis index decreased in patients with chronic gingivitis consuming lettuce juice regularly during 2 weeks12,14. In our study, the addition of 5 mM nitrate led to a significant decrease of Porphyromonas gingivalis, Treponema maltophilum (both strongly associated with periodontitis), and an unclassified Streptococcus sp. Streptococcus spp. are generally periodontal health-associated (with the exception of S. constellatus), but decrease in the presence of nitrate in healthy individuals, possibly because nitrate could prevent the overgrowth of aciduric representatives16. The 50 mM nitrate condition increased an unclassified Aggregatibacter species, an unclassified Neisseria species and Rothia mucilaginosa, while decreasing 25 species of which most were associated with periodontitis on a genus- or species-level. Rothia and Neisseria spp. are considered periodontal-health associated21, while Aggregatibacter is often associated with periodontitis, because of the strong association of A. actinomycetemcomitans with periodontitis in some populations28. However, this genus can also contain species that are not disease-associated. Regarding this, A. actinomycetemcomitans was detected as a classified species and did not change significantly in the presence of nitrate. The classified species that significantly decreased in the presence of 50 mM nitrate included Fusobacterium nucleatum, Dialister invisus, Tannerella forsythia, Alloprevotella tannerae, Eikenella corrodens, Filifactor alocis, Porphyromonas endodontalis and Prevotella intermedia, which are strongly associated with periodontitis10,12,29 and contributed to a reduced dysbiosis index.
The observation that 50 mM nitrate caused a larger decrease in dysbiosis index than 5 mM nitrate, indicates that the topical application of nitrate to achieve this concentration should be tested in future studies. It should be noted that 100 mM nitrate (without further reduction to nitrite and other compounds) can be toxic for oral species26 and it is unknown if the 50 mM nitrate used in our experiment directly inhibited certain species (i.e. exhibiting an antiseptic or toxic effect on specific species instead of prebiotic effect that shifts to community towards eubiosis). Regarding this, some species that decreased in the presence of 50 mM nitrate (e.g., Capnocytophaga leadbetteri and Granulicatella elegans) were not periodontitis-associated. Nitrite and nitric oxide are often toxic for bacteria at lower doses than nitrate (e.g., S. mutans was inhibited by 100 mM nitrate but by 0.5 mM nitrite)26. The reduction of nitrate to nitrite and further metabolization to nitric oxide in our in vitro conditions could also explain the decrease of different species in the 5 mM and 50 mM nitrate conditions. For example, the strongly periodontitis-associated Porphyromonas gingivalis can be killed by nitric oxide17 and decreased in both conditions.
The concentration of 5 mM nitrate can be obtained in saliva by vegetable consumption, which is considered a safe and recommendable strategy to increase the nitrate levels inside the oral cavity16. To obtain higher levels of nitrate, topical applications could be used such as toothpaste or, for the application in periodontal pockets, a periodontal gel. The safety of higher levels of nitrate and nitrate-reducing probiotics has been discussed previously in Rosier et al.16 and 2020b24, respectively. In short, doses far below the acceptable daily intake (3.7 mg nitrate/kg of body weight, which is 222 mg for an adult of 60 kg) could be added to oral hygiene products to obtain high concentrations of nitrate. For example, a concentration of 50 mM can be obtained by adding 3.1 mg of nitrate to a dose of 1 ml periodontal gel. Additionally, nitrate-rich vegetable extracts could be used containing natural antioxidants, or nitrate salts could be combined with antioxidants (e.g., vitamin C), to prevent potential N-nitroso compound formation. This could be especially relevant for supragingival biofilms in which the pH can reach low levels, stimulating N-nitroso compound formation16, while in periodontal pockets the pH tends to be neutral or slightly alkaline. The possibility of N-nitroso compound formation and the inhibition with antioxidants should be investigated in future studies.
The Ra9 strain in combination with 5 mM nitrate resulted in more significant changes (20 in total) on a species-level compared with just 5 mM nitrate (3 significant changes). These included periodontitis-associated species (e.g., Selenomonas sputigena, Eikenella corrodens and Fusobacterium nucleatum), but also health-associated and (potentially) nitrate-reducing species (e.g., Corynebacterium matruchotii and Rothia mucilaginosa). The decrease in a variety of species could have resulted from nitric oxide produced by Ra9, which contains nitrite-reduction genes24 and/or other potential antimicrobial mechanisms of this bacterium. Genes of Rothia species involved in the production of antimicrobial peptides, enterobactin (a metal chelating siderophore with antimicrobial activity) and valinomycin (an antimicrobial ionophore) have been identified30, and the possibility of such mechanisms affecting the oral microbiota should be explored. Additionally, Ra9 could have competed with nutrients and other compounds (e.g., nitrate and lactic acid as electron donor) with species with a similar type of metabolism, such as Rothia mucilaginosa. Both Rothia mucilaginosa and Corynebacterium matruchotii are among the health-associated species that affect the dysbiosis index. This could explain why the mean dysbiosis index of communities treated with Ra9 and 5 mM nitrate did not decrease significantly. It should be noted that a small effect on pH was observed (i.e., after the 7 h incubation period, the supernatant was pH 6.3 when adding Ra9 and pH 6.5 when adding Ra9 in combination with nitrate compared with pH 6.7 of the control condition, both p < 0.05), possibly resulting from the metabolism of the added Ra9 strain or from changes in the activity of other members of the bacterial community. In future studies, pH decreases introduced by probiotics (e.g., the currently used acidogenic Lactobacillus spp.31 that are likely to induce a larger pH drop) should be investigated as they could affect alkalophilic periodontitis-associated species or (in some cases) stimulate enamel demineralization when pH ~5.5 is reached. Our data indicates that the effect of Ra9 on bacterium composition and inflammation should be further explored in vivo as potential pro-, post- or synbiotic treatment. A limitation of our in vitro model is that there is no interaction with human host cells, which respond to changes in microbial composition and metabolites, including nitric oxide32. Based on a limited amount of clinical studies focusing on oral microbiota composition, inflammatory cytokines and/or clinical parameters, both nitrate15 and Rothia spp. are associated with reduced gingival inflammation14,18,20,33.
A consistent observation in our study was the decrease of Porphyromonas gingivalis or other Porphyromonas species in the presence of nitrate (with the exception of 1 unclassified Porphyromonas species that increased slightly when treating the communities with Ra9 and 5 mM nitrate). Additionally, Fusobacterium nucleatum and an unclassified Fusobacterium species decreased, which is consistent with P. gingivalis and F. nucleatum being sensitive to the oxidative stress of nitric oxide16. To test the effect of nitrate on these species, an established 10-species biofilm model was used, including these two species and two nitrate-reducing species (Veillonella dispar and Actinomyces naeslundii). In this model, the bacteria are grown for 1 week to form a biofilm and the species are quantified by qPCR. The addition of 6.5 mM nitrate led to a significant decrease in Porphyromonas gingivalis, Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans. These three species are strongly associated with periodontitis, while two of them (P. gingivalis and F. nucleatum) are also associated with halitosis, and their decrease is undoubtedly a beneficial change for oral health. This further supports the idea that the unclassified Porphyromonas and Aggregatibacter species that increased in different nitrate conditions are not related to these periopathogens. In line with our results, in a previous study using saliva of healthy individuals, 6.5 mM nitrate also led to a decrease of Porphyromonas and Fusobacterium on a genus level. However, these results have not been confirmed in clinical studies in which nitrate intake increased Rothia and/or Neisseria, while it decreased Streptococcus, Veillonella and/or Prevotella34,35 (reviewed by Rosier et al.16). Larger clinical studies in which changes are monitored on a species- and strain-level are needed to conclude the effects of nitrate on the oral microbiota in individuals with and without oral diseases.
When adding Ra9 in combination with 5 mM nitrate, more nitrate was reduced, and more nitrite was produced. This could be very relevant for human health, as nitrite production could improve systemic conditions such as hypertension, metabolic syndrome and diabetes, and can increase endothelial function and sport performance36. Interestingly, periodontitis, which is unequivocally linked to lower levels of nitrate-reducing organisms in subgingival plaque, has been associated with an increased risk of these systemic conditions37. Additionally, preeclampsia is one of the conditions associated with periodontitis and a recent study indicated that this condition was associated with a decrease in oral nitrate-reducing bacteria38. Therefore, the relationship between periodontitis, nitrate reduction and systemic complications and diseases should be investigated in future studies, as lower levels of nitrate-reducing bacteria in periodontitis patients could lead to a decrease in systemic nitric oxide levels, increasing the risk of different systemic conditions. We propose that nitrate-reducing probiotics could therefore increase nitric oxide availability, thereby reducing the risk of systemic conditions and complications that are stimulated by a nitric oxide deficit. Additionally, it should be explored if nitrate-reducing probiotics could improve sport performance. Interestingly, nitrate has clear anti-caries effects39, while athletes have an increased caries prevalence due to high carbohydrate consumption40. Nitrate with or without nitrate-reducing bacteria as a prebiotic or synbiotic treatments should be further explored in athletes to improve sport performance and oral health.
In conclusion, our data indicates that nitrate and nitrate-reducing bacteria should be explored as pre-, pro- and synbiotic treatments for periodontitis and the systemic complications associated with this oral disease. We found that nitrate can reduce biofilm growth and periodontitis-associated species in dysbiotic communities from periodontal pockets.
Methods
Bacterial strains and growth conditions
Rothia aeria CECT9999 (Ra9, isolate D1P7 described in24), was used as a nitrate-reducing probiotic candidate. Prior to the experiment, Brain Heart Infusion (BHI) (Biolife, Deerfield, Illinois, USA) agar plates were inoculated with the strain, incubated for 48 h at 37 °C, and stored at 4 °C for up to 14 days. A single colony was transferred from the BHI agar plates to liquid BHI and incubated for 24–48 h at 37 °C before inoculation to the biofilm culture.
Patient inclusion and sampling procedure
Bacterial samples from periodontal pockets were collected at the Lluis Alcanyis Foundation dental clinic of the University of Valencia (Valencia, Spain) and the private clinic Centro Periodontal de Valencia (Valencia, Spain). The study protocol was approved by the Ethics Committee of the University of Valencia (Spain) (H1547805836517) and all participants signed an informed consent prior to sample donation.
Thirty-one periodontitis patients were recruited at the clinics before starting treatment. The included patients were ≥18 years of age and had a sufficient number of teeth in each quadrant (≥5). An overview of patient characteristics is included in Supplemental Table 1. The diagnosis of periodontitis was determined in accordance with the guidelines of The American Academy of Periodontology; (AAP)41. Exclusion criteria were (i) the use of systemic antibiotics or antiseptic mouthwash in the last month, (ii) active systemic infection, (iii) periodontal treatment in the previous 6 months, (iv) and pregnancy. The patients included were 16 males and 14 females, leading to a total of 30 individuals of which 11 were smokers. The patients’ age ranged from 20–76, with a mean age of 49 ± 14. Their average deepest pocket depth was 6.4 ± 1.9 mm.
Subgingival plaque was collected from the periodontal pocket at five different periodontitis sites in each patient using 6–10 sterile paper points. Paper points were transferred to a 2 mL tube containing BHI medium (for usage within 1 h) or reduced transport medium42 (for usage within 1–12 h) and stored at ~4 °C in a styrofoam box for transportation. The samples were transported to the laboratory and tested within 1–12 h.
Real-time in vitro biofilm growth of periodontal plaque
Real-time in vitro biofilm growth of oral microorganisms was assessed using the xCELLigence RTCA single plate system (ACEA Biosciences)13,43. When grown in this system, bacteria from fresh subgingival samples attach to the wells interfering with the electrical current of the electrodes, and the resulting impedance allows the real-time quantification of biofilm growth22. The amount of biofilm growth is measured and expressed as “Cell Index” values, which correlate with total biofilm mass44.
Baseline cell-sensor impedance measurements were performed with 100 µl liquid Brain Heart Infusion broth (BHI) medium (Biolife) supplemented with 2 mL/L liquid vitamin K1 (V3501), 5 mg/L hemin and 1 mg/L menadione (all Sigma-Aldrich, St. Louis, Missouri, USA). Three baseline measurements of the control medium were performed in a 96-well plate with an integrated microelectronic cell sensor array (E-plate 96, ACEA Biosciences, San Diego, California, USA) at 3-min intervals. After this, another 50 µl BHI medium was added, which was supplemented to obtain the following treatment groups: 5 mM nitrate (Thermo Fisher Scientific), 50 mM nitrate, 5 mM nitrate combined with the Ra9 strain (OD600nm Ra9 in well = 0.075), or the Ra9 strain alone (OD600nm Ra9 in well = 0.075) (see Table 1 for details). Finally, 100 µl of BHI medium containing periodontal plaque as inoculum was added from an individual bacterial inoculum sample per patient. This bacterial inoculum was prepared by vortexing the paper points for 1 min, discarding the paper points, centrifuging the periodontal plaque sample (1 min, 9660 xg), discarding the (transport) medium supernatant, and re-suspending the bacterial pellet in BHI medium (100 µl per paper point, which is 600–1000 µl for 6–10 paper points). Replicates of each condition were prepared for every patient.
Concisely, the subgingival plaque samples were grown in BHI with or without 5 mM nitrate medium (n = 9), 50 mM nitrate medium (n = 10), or 5 mM nitrate, 5 mM nitrate + Ra9, and Ra9 medium (n = 11) (Table 1). All plates were sealed with adhesive aluminium foil and incubated at 37 °C. Cell impedance measurements were performed at 10-min intervals. The total growth period was 7 h when using 5 mM nitrate (the same time as used previously for healthy communities by Rosier et al.,24) or 12 h for 50 mM nitrate to obtain a ~50% of nitrate reduction of each concentration. Supernatants, as well as the corresponding biofilms collected through resuspension using PBS, were collected from the 96-well plate after the final cell-sensor impedance measurement. All samples were stored at −20 °C.
Determination of pH, nitrate, and nitrite concentrations
Supernatant collected from the in vitro biofilms was used to determine the pH levels and the concentration of nitrate and nitrite using the RQflex® 10 Reflectoquant® reflectometer (Merck KGaA), following Rosier et al.13. Manufacturer protocols were adjusted for micro-volume sample sizes, and verified using known concentrations. The supernatant was applied undiluted to pH test strips (reference no. 1169960001) or as a ≥ 10-fold dilution of supernatant in demineralized water to the nitrate (reference no. 1169710001) and the nitrite (reference no. 1169730001) test strips. Fifteen μl of the sample was directly pipetted onto each of the two reaction zones of a test strip, excess liquid was removed by tipping the side of the strip on a tissue, and the strip was incubated according to the manufacturer’s instructions.
DNA isolation
Bacterial DNA was extracted for Illumina sequencing of the 16 S rRNA gene. Bacterial pellets (two replicates of biofilms after in vitro growth or the pellet of 100 μl inoculum) were resuspended in 100 μl PBS and sonicated in a RAYPA ultrasonic bath for 30 s. To lyse the samples, they were incubated with lysozyme (20 mg/ml; Thermomixer comfort, Eppendorf), lysostaphin (2000 units/mg protein; Sigma-Aldrich) and mutanolysin (4000 units/mg protein; Sigma-Aldrich) for 60 min at 37 °C13. Next, DNA was extracted by the MagNA Pure LC 2.0 Instrument (Roche Diagnostics, Barcelona, Spain) with the MagNA Pure LC DNA Isolation Kit III for Bacteria and Fungi (Roche Diagnostics GmbH) using standard manufacturer protocol. DNA was eluted in 100 μl elution buffer and, if necessary (e.g., for some inoculum samples), further concentrated with a Vivacon® 500 100.000 MWCO Hydrosart filter (Sartorius). A QubitTM 3 Fluorometer (ThermoScientific) was used to determine DNA concentration and samples were stored at −20 °C until used for sequencing.
Illumina 16 S rRNA gene sequencing and analysis
DNA from biofilms and inocula were sequenced to determine the bacterial composition. Library preparation was performed according to the 16 S rDNA gene Metagenomic Sequencing Library Preparation Illumina protocol (Part #15044223 Rev. A, Illumina Inc.), using gene-specific 16 S amplicon PCR primers for the V3 and V4 region, resulting in amplicons of ~460 bp (F: 5' TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNG GCWGCAG 3'; R: 5' GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC 3'). The resulting library was sequenced with an Illumina MiSeq instrument using the 2 × 300 bp paired-ends protocol following the manufacturer’s instructions.
To process the paired-end FASTQ files, an amplicon sequence variant (ASV) table was obtained using the DADA2 pipeline (v1.8) in R45 following Rosier et al.,39. In summary, the forward and reverse reads were trimmed, removing the primer sequences and low-quality bases at the end of the reads through end-trimming. Reads with any ambiguous N base or exceeding 5 expected errors were also discarded. The forward and reverse pairs were merged, with a minimum overlap of 12 bases and a maximum mismatch of 1 base in the overlapping region, to obtain the single denoised variants. After chimera removal, the final amplicon sequence variants (ASVs) were mapped onto the Homo sapiens genome (assembly GRCh38.p13), using Bowtie246 (v2.3.5.1), in order to remove artefactual reads from the host. The Silva database47,48 (v138) was set as a reference to assign taxonomy to each ASV. Genus classification was achieved using the DADA2 naive Bayesian classifier method. The ASVs with an assigned genus but without exact species, were aligned using the Blastn tool49 (v2.10.0+) against the Silva database with a minimum of 97% of identity.
The absolute and normalized abundance of bacterial genera and species could be computed for graphing and further analysis. Bacterial abundance was normalized using ANCOM-BC approach to account for the compositional nature of 16 S data50.
SMDI score
The Subgingival Microbial Dysbiosis Index (SMDI) was computed for each treatment condition as described by Chen et al.12. In short, read counts tables were generated at the species level and normalized by centered log-ratio (CLR) transformation. Based on a list of discriminating species (DS) compiled at different cut-offs of mean decrease in Gini (MDG), which is a measure of the importance of a taxon to the classification, and their mean CLR abundances in health and periodontitis as determined in the study by Chen et al.12, a SMDI score was calculated for each inoculum or biofilm sample as follows: SMDI = mean CLR abundance of dysbiotic DS—mean CLR abundance of “normobiotic” (i.e., the composition in health) DS.
10-species biofilm model and nitrate
To test the effect of nitrate on oral bacteria, a multi-species in vitro biofilm model containing a synthetic community of 10 oral strains (Streptococcus mitis NCTC 12261, S. intermedius DSM 20753, S. oralis NTCC 11427, Fusobacterium nucleatum ATCC 10596, F. nucleatum spp. vincentii DSM 19507, Actinomyces naeslundii DSM 17233, Veillonella dispar NCTC 11831, Porphyromonas gingivalis W83, Prevotella intermedia DSM 20706, and Aggregatibacter actinomycetemcomitans) was used as described by Brown et al. with a nitrate condition. In short, the in vitro biofilms were grown with or without 6.5 mM nitrate for 7 days23. A qPCR was performed to quantify the levels of the 10 bacterial species using previously published primer sets23. These primers were used to detect Streptococcus spp. (F: 5' GATACATAGCCGACCTGAG 3'; R: 5' TCCATTGCCGAAGATTCC 3'), A. naeslundii (F: 5' GGCTGCGATACCGTGAGG 3'; R: 5' TCTGCGATTACTAGCGACTCC 3'), V. dispar (F: 5' CCGTGATGGGATGGAAACTGC 3'; R: 5' CCTTCGCCACTGGTGTTCTTC 3'), Fusobacterium spp. (F: 5' GGATTTATTGGGCGTAAAGC 3'; R: 5' GGCATTCCTACAAATATCTACGAA 3'), P. gingivalis (F: 5' GCGCTCAACGTTCAGCC 3'; R: 5' CACGAATTCCGCCTGC 3'), P. intermedia (F: 5' CGGTCTGTTAAGCGTGTTGTG 3'; R: 5' CACCATGAATTCCGCATACG 3'), and A. actinomycetemcomitans (F: 5’ GAACCTTACCTACTCTTGACATCCGAA 3’; R: 5' TGCAGCACCTGTCTCAAAGC 3').
Statistical analysis
Statistical analysis of real-time in vitro biofilm growth of periodontal plaque, determination of pH, nitrate, and nitrite in the supernatants, SMDI score and Shannon diversity of the biofilm composition, and qPCR quantification of the 10-species biofilm model was performed using a nonparametric Wilcoxon matched-pairs signed rank test with Bonferroni multiple testing correction for multiple comparisons using IBM SPSS statistics (version 27), and considered statistically significant at a two-sided p-value < 0.05. The qPCR data of the 10-species biofilm model was analyzed using a Mann–Whitney U-test with Bonferroni–Dunn multiple testing correction.
Fold changes in microbiome composition were tested using ANCOM-BC, and considered statistically significant at an adjusted p-value < 0.0550. Boruta feature selection51 of species identified as significantly different between treatment conditions by ANCOM-BC was applied to further highlight relevant changes. In this study only genera with a median abundance >0.1% were discussed. Data were visualized using GraphPad Prism 8 (version 8.3.0). Other statistical analyses of the bacterial microbiome data were performed according to Johnston et al. using R9. Shannon diversity, Adonis tests (Permutational Multivariate Analysis of Variance Using Distance Matrices) and visualization of bacterial composition in a two-dimensional map using constrained correspondence analysis (CCA) were performed using the R Vegan library52.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Sequencing data is available in the NCBI sequence read archive (SRA) under BioProject PRJNA951622. All other data generated or analyzed during this study are included in this published article and its Supplementary Information file or are available from the corresponding author upon reasonable request.
References
Marsh, P. D. Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294 (2003).
Rosier, B. T., Marsh, P. D. & Mira, A. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J. Dent. Res. 97, 371–380 (2018).
Albandar, J. M., Streckfus, C. F., Adesanya, M. R. & Winn, D. M. Cigar, pipe, and cigarette smoking as risk factors for periodontal disease and tooth loss. J. Periodontol. 71, 1874–1881 (2000).
Kassebaum, N. J. et al. Global burden of severe periodontitis in 1990-2010: a systematic review and meta-regression. J. Dent. Res. 93, 1045–1053 (2014).
Preshaw, P. M. et al. Periodontitis and diabetes: a two-way relationship. Diabetologia 55, 21–31 (2012).
Chou, Y. Y., Lai, K. L., Chen, D. Y., Lin, C. H. & Chen, H. H. Rheumatoid arthritis risk associated with periodontitis exposure: a nationwide, population-based cohort study. PLoS One 10, e0139693 (2015).
Kebschull, M., Demmer, R. T. & Papapanou, P. N. “Gum bug, leave my heart alone!”-epidemiologic and mechanistic evidence linking periodontal infections and atherosclerosis. J. Dent. Res. 89, 879–902 (2010).
Macedo Paizan, M. L. & Vilela-Martin, J. F. Is there an association between periodontitis and hypertension? Curr. Cardiol. Rev. 10, 355–361 (2014).
Johnston, W. et al. Mechanical biofilm disruption causes microbial and immunological shifts in periodontitis patients. Sci. Rep. 11, 1–14 (2021).
Pérez-Chaparro, P. J. et al. Newly identified pathogens associated with periodontitis: a systematic review. J. Dent. Res. 93, 846–858 (2014).
Darveau, R. P. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8, 481–490 (2010).
Chen, T., Marsh, P. D. & Al-Hebshi, N. N. SMDI: An index for measuring subgingival microbial dysbiosis. J. Dent. Res. 101, 331–338 (2022).
Rosier, B. T., Buetas, E., Moya-Gonzalvez, E. M., Artacho, A. & Mira, A. Nitrate as a potential prebiotic for the oral microbiome. Sci. Rep. 10, 12895 (2020).
Jockel-Schneider, Y. et al. A nitrate-rich diet alters the composition of the oral microbiota in periodontal recall patients. J. Periodontol. https://doi.org/10.1002/JPER.1020-0778 (2021).
Jockel-Schneider, Y. et al. Stimulation of the nitrate-nitrite-NO-metabolism by repeated lettuce juice consumption decreases gingival inflammation in periodontal recall patients: a randomized, double-blinded, placebo-controlled clinical trial. J. Clin. Periodontol. 43, 603–608 (2016).
Rosier, B. T. et al. The importance of Nitrate reduction for oral health. J. Dent. Res. 101, 887–897 (2022).
Backlund, C. J., Sergesketter, A. R., Offenbacher, S. & Schoenfisch, M. H. Antibacterial efficacy of exogenous nitric oxide on periodontal pathogens. J. Dent. Res. 93, 1089–1094 (2014).
Corrêa, J. D. et al. Oral microbial dysbiosis linked to worsened periodontal condition in rheumatoid arthritis patients. Sci. Rep. 9, 8379 (2019).
Joshi, V. et al. Smoking decreases structural and functional resilience in the subgingival ecosystem. J. Clin. Periodontol. 41, 1037–1047 (2014).
Huang, S. et al. Longitudinal multi-omics and microbiome meta-analysis identify an asymptomatic gingival state that links gingivitis, periodontitis, and aging. mBio 12, e03281–03220 (2021).
Feres, M., Retamal-Valdes, B., Gonçalves, C., Cristina Figueiredo, L. & Teles, F. Did Omics change periodontal therapy? Periodontol. 2000. 85, 182–209 (2021).
Mira, A. et al. Development of an in vitro system to study oral biofilms in real time through impedance technology: validation and potential applications. J. Oral. Microbiol. 11, 1609838 (2019).
Brown, J. L. et al. Biofilm-stimulated epithelium modulates the inflammatory responses in co-cultured immune cells. Sci. Rep. 9, 15779 (2019).
Rosier, B. T., Moya-Gonzalvez, E. M., Corell-Escuin, P. & Mira, A. Isolation and characterization of Nitrate-reducing bacteria as potential probiotics for oral and systemic health. Front. Microbiol. 11, 555465 (2020).
Takahashi, N. Microbial ecosystem in the oral cavity: metabolic diversity in an ecological niche and its relationship with oral diseases. Int. Congr. Ser. 1284, 103–112 (2005).
Wicaksono, D. P., Washio, J., Abiko, Y., Domon, H. & Takahashi, N. Nitrite production from Nitrate and its link with lactate metabolism in oral veillonella spp. Appl. Environ. Microbiol. 86, e01255–01220 (2021).
Cosseau, C. et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect. Immun. 76, 4163–4175 (2008).
Haubek, D. et al. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet 371, 237–242 (2008).
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C. & Kent, R. L. Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 (1998).
Oliveira, I. M. F. et al. Comparative genomics of Rothia species reveals diversity in novel biosynthetic gene clusters and ecological adaptation to different eukaryotic hosts and host niches. Microbial Genom. 8, mgen000854 (2022).
Amato, M. et al. Probiotics in periodontal and peri-implant health management: biofilm control, dysbiosis reversal, and host modulation. Microorganisms 10, 2289 (2022).
Hezel, M. P. & Weitzberg, E. The oral microbiome and nitric oxide homoeostasis. Oral Dis. 21, 7–16 (2015).
Khocht, A. et al. Cross-sectional comparisons of subgingival microbiome and gingival fluid inflammatory cytokines in periodontally healthy vegetarians versus non-vegetarians. J. Periodontal Res. 56, 1079–1090 (2021).
Burleigh, M. et al. Dietary nitrate supplementation alters the oral microbiome but does not improve the vascular responses to an acute nitrate dose. Nitric Oxide 89, 54–63 (2019).
Vanhatalo, A. et al. Nitrate-responsive oral microbiome modulates nitric oxide homeostasis and blood pressure in humans. Free Radic. Biol. Med. 124, 21–30 (2018).
Lundberg, J. O., Carlström, M. & Weitzberg, E. Metabolic effects of dietary nitrate in health and disease. Cell Metab. 28, 9–22 (2018).
Martínez-García, M. & Hernández-Lemus, E. Periodontal inflammation and systemic diseases: an overview. Front. Physiol. 12, 709438 (2021).
Altemani, F., Barrett, H. L., Callaway, L. K., McIntyre, H. D. & Dekker Nitert, M. Reduced abundance of nitrate-reducing bacteria in the oral microbiota of women with future preeclampsia. Nutrients 14, 1139 (2022).
Rosier, B. T. et al. A single dose of nitrate increases resilience against acidification derived from sugar fermentation by the oral microbiome. Front. Cell Infect. 11, 692883 (2021).
Burleigh, M. C., Sculthorpe, N., Henriquez, F. L. & Easton, C. Nitrate-rich beetroot juice offsets salivary acidity following carbohydrate ingestion before and after endurance exercise in healthy male runners. Plos One 15, e0243755 (2020).
AAP. The American Academy of Periodontology 2000 annual report. J. Periodontol. 71, 1943–1957 (2000).
Syed, S. A. & Loesche, W. J. Survival of human dental plaque flora in various transport media. Appl. Microbiol. 24, 638–644 (1972).
Ferrer, M. D., Lamarche, B. & Mira, M. Studying Bacterial Biofilms Using Cellular Impedance. J. Appl. Microbiol. 122, 640–650 (2016).
Ferrer, M. D. et al. Effect of antibiotics on biofilm inhibition and induction measured by real-time cell analysis. J. Appl. Microbiol. 122, 640–650 (2016).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Yilmaz, P. et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648 (2014).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Lin, H. & Peddada, S. D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514 (2020).
Kursa, M. B. & Rudnicki, W. R. Feature selection with the Boruta package. J. Stat. Softw. 36, 1–13 (2010).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.4-2. http://CRAN.R-project.org/package=vegan (2017).
Acknowledgements
We would like to thank the staff and patients of the Lluis Alcanyis Foundation dental clinic of the University of Valencia (Valencia, Spain) and the private clinic Centro Periodontal de Valencia (Valencia, Spain) for their help and participation in this study. We are also very grateful to Dr. Tsute Chen (Department of Microbiology, Forsyth Institute, Cambridge, United States of America) for his help with the SMDI score analysis and to Prof. Gordon Ramage (Oral Sciences Research Group, Glasgow Dental School, School of Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, Scotland) for his help with setting up the 10-species biofilm model. A.M. was supported by a grant from the European Regional Development Fund and Spanish Ministry of Science, Innovation and Universities with the reference RTI2018-102032-B-I00, as well as a grant from the Valencian Innovation Agency with the reference INNVAL20/19/006. BR was supported by a FPI fellowship from the Spanish Ministry of Science, Innovation and Universities with the reference Bio2015-68711-R.
Author information
Authors and Affiliations
Contributions
B.T.R. and A.M. contributed to the design of work, and B.T.R., A.M. and D.M. drafted and revised the manuscript. C.L., C.P.M. and F.A. performed the subgingival plaque sample collection D.M. and T.L. did most experimental work and M.C.-D. did most bioinformatic work. All authors contributed to data the acquisition and analysis, and all authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: A.M. and B.T. Rosier are coinventors in a pending patent application owned by the FISABIO Institute, which protects the use of nitrate as a prebiotic and certain nitrate-reducing bacteria as probiotics. The remaining authors declare no competing interests.
Ethics approval and consent to participate
All donors gave informed consent prior to sample collection. The study protocol was reviewed and approved by the Ethics Committee of the University of Valencia (Spain) (H1547805836517). This study was carried out according to the relevant guidelines and regulations of the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mazurel, D., Carda-Diéguez, M., Langenburg, T. et al. Nitrate and a nitrate-reducing Rothia aeria strain as potential prebiotic or synbiotic treatments for periodontitis. npj Biofilms Microbiomes 9, 40 (2023). https://doi.org/10.1038/s41522-023-00406-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-023-00406-3
- Springer Nature Limited
This article is cited by
-
Nitrate reduction capacity of the oral microbiota is impaired in periodontitis: potential implications for systemic nitric oxide availability
International Journal of Oral Science (2024)
-
Evaluating the Role of Postbiotics in the Modulation of Human Oral Microbiota: A Randomized Controlled Clinical Trial
Probiotics and Antimicrobial Proteins (2024)