Abstract
Eastern Equine Encephalitis virus (EEEV) is an alphavirus that can cause severe diseases in infected humans. The very low-density lipoprotein receptor (VLDLR) was recently identified as a receptor of EEEV. Herein, we performed cryo-electron microscopy structural and biochemistry studies on the specific interactions between EEEV and VLDLR. Our results show that VLDLR binds EEEV at three different sites A, B and C through its membrane-distal LDLR class A (LA) repeats. Site A is located in the cleft in between the E1-E2 heterodimers. Site B is located near the connecting β ribbon of E2 and is in proximity to site A, while site C is on the domain B of E2. The binding of VLDLR LAs to EEEV is in complex modes, including the LA1-2 and LA3-5 mediated two major modes. Disruption of the LA1-2 mediated binding significantly affect the cell attachment of EEEV. However, the mutation W132G of VLDLR impairs the binding of LA3, drives the switch of the binding modes, and significantly enhances the attachment of EEEV to the cell. The W132G variant of VLDLR could be identified in human genome and SNP sequences, implying that people with similar mutations in VLDLR may be highly susceptible to EEEV infection.
Similar content being viewed by others
Introduction
The Eastern equine encephalitis virus (EEEV) is an alphavirus that can cause severe and sometimes fatal disease in humans and equines1,2. EEEV is commonly found in the eastern regions of America and has been associated with outbreaks of encephalitis3. Birds are the major enzootic hosts and natural reservoirs of EEEV. EEEV is transmitted among birds mainly through the mosquito Culiseta melanura4. Other mosquito species, like Aedes and Coquillettidia, can also transmit EEEV and are believed to be the bridge vectors for the transmission of EEEV from birds to humans and horses4,5. Unlike the encephalitic alphavirus Venezuelan equine encephalitis virus (VEEV) efficiently amplifies and develops a high viremia in infected equines, EEEV infection of humans or horses can not develop a high viremia that is sufficient for establishing a complete transmission cycle6,7. Antigenic variants of EEEV are categorized into two major groups, including the more virulent North American variant and the less virulent South American Variant8. So far, there is still no available vaccine or drug for the prevention and treatment of EEEV infection in humans.
The spherical virions of EEEV are approximately 70 nm in diameter and are assembled by the viral RNA genome, the envelope proteins E1, E2, the capsid protein C, and the small 6 kDa protein 6 K. A transframe protein TF could also be generated from the 6 K gene by −1 ribosomal frameshifting during translation9,10,11,12. The envelope and capsid proteins of EEEV assemble to form two concentric protein shells that are separated by a lipid bilayer. The envelope protein shell is composed of 80 trimeric spikes that are arranged with T = 4 icosahedral symmetry13,14,15,16. Twenty of the 80 spikes are sitting on the icosahedral 3 fold axes (i3) and sixty of the 80 spike are sitting on the quasi 3 fold axes (q3). Each trimeric spike is an assembly of three E1 and E2 heterodimers and can be divided into two distinct parts, including a flower-like protruding portion and a shell-forming portion. The flower-like protruding portion is composed by the three immunoglobulin-like β domains A (residues 1-132), B (residues 173-231), and C (residues 269-342) of E2 and the immunoglobulin-like β domain 2 (residues 40-127 and 170-272) of E1 (E1-DII)10,11,16. Domains A and C of E2s are located in the center of the spike and are surrounded by the E1-DIIs. The three E2 domain Bs are arranged anti-clockwisely on the top of the spike and constitute three petals of the flower. The B domain of E2 is connected to the domains A and C of E2 through two long linker peptide (residues 133-172, 231-268) named the “β-ribbon connector”. The E1-DI (residues 1-36, 133-168, 274-293) and DIII (residues 294-381) at the base of the spike splay tangentially along the viral surface and interact with each other to constitute a smooth protein shell16.
Like other alphaviruses, EEEV enters host cells through receptor-mediated endocytosis. The spikes of the E1-E2 heterodimers are responsible for specific receptor attachment10,11. Recently, the very low-density lipoprotein receptor (VLDLR) and the apolipoprotein E receptor 2 (ApoER2), two closely related receptors from the low-density lipoprotein (LDL) receptor family, were identified as the receptors of EEEV17. LDLRAD3 of the same LDL receptor family was identified as an entry receptor of VEEV18. VLDLR and ApoER2 can also mediate the entry of Semliki Forest virus (SFV) and Sindbis virus (SINV), two alphaviruses that rarely cause encephalitis in equines or humans17. The Matrix Remodeling Associated protein 8 (MXRA8) was identified as an entry receptor of many other alphaviruses19,20.
Receptors from the LDL receptor family have similar small LDLR class A (LA) repeats in their ectodomains21. The small LA repeat domain has approximately 40 amino acid residues and a compact fold that contains a calcium-binding motif and is crosslinked by three disulfide bonds21,22,23,24. Although both VLDLR and LDLRAD3 bind the viruses through their ecto small LA repeat domains, these two receptors bind the viruses with distinct different modes. LDLRAD3 binds through its distal LA repeat domain in clefts between the E1-E2 heterodimers of the protruding VEEV spikes25,26. However, VLDLR binds SFV on the smooth shell constituted by E1-DIIIs in a synergistic mode that involves multiple ecto LA repeat domains of the receptor27. Comparing to VLDLR and LDLRAD3, MXRA8 has a quite different structure and binds the viruses through its distal Ig-like domains. Of note, MXRA8 bind in clefts between the E1-E2 heterodimers of the protruding spikes as does LDLRAD3, although the interactions are completely different19,28,29. Our recent results showed that it is unlikely that VLDLR binds EEEV in a similar mode as does SFV, which was confirmed by a recent study30. The structural basis for the selective binding of VLDLR to EEEV need to be further investigated.
In this work, we report cryo-EM structures of EEEV in complex with VLDLR. We found that VLDLR uses its membrane-distal LA repeats to bind EEEV at three different sites A, B, and C on the surface trimeric spikes. LA1 and LA5 specifically recognize site A, whereas LA2, LA3, and LA6 can bind both sites A and C. Combined with biochemistry studies, we identified multiple different binding modes of VLDLR, including two major binding modes mediated by LA1-2 and LA3-5. Our results provide new insights into the receptor-mediated entry of alphaviruses.
Results
Structure of VLDLR LA1-8 in complex with EEEV
The membrane-distal homologous LDLR class A (LA) repeats of VLDLR and ApoER2 were shown to mediate the interactions between EEEV and VLDLR17. ApoER2 have two isoforms and the isoform2 (ApoER2iso2) is the prodominent form. VLDLR contains eight LA repeats (VLDLR LA1-8), while ApoER2iso2 contains three LA repeats (ApoER2iso2 LA1-3). Sequence alignments show that the sequence identity between LA1-3 of VLDLR and LA1-3 of ApoER2iso2 is 55.4% (Supplementary Fig. 1a). We, thus, produced VLDLR LA1-8 and ApoER2iso2 LA1-3 in fused with an antibody constant region fragment at the C-terminus, and prepared the complexes with the EEEV virus-like particles (VLPs), which were produced by using the structural proteins of the EEEV PE6 strain31. The structures of the complexes were determined by using cryo-electron microscopy (cryo-EM) single-particle reconstruction at a resolution of 5.3 Å for the VLDLR LA1-8-Fc and EEEV PE6 VLP complex and 4.9 Å for the ApoER2iso2 LA1-3 and EEEV PE6 VLP complex with icosahedral symmetry imposed (Supplementary Fig. 1b–f, Supplementary Table 1). Compared to the LA free EEEV PE6 VLPs, we identified additional densities on the spikes of VLDLR LA1-8-Fc-EEEV PE6. However, we did not observe additional density in the reconstruction of ApoER2iso2 LA1-3-EEEV PE6 (Supplementary Fig. 2a). Further block-based reconstructions with the asymmetric unit as a block were calculated for the VLDLR LA1-8-Fc and EEEV PE6 VLP complex and yield a density map at a resolution of 3.5 Å (Supplementary Fig. 1b–e).
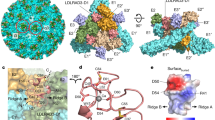
Additional densities at three distinct sites A, B, and C were observed on the spikes of VLDLR LA1-8-Fc-EEEV PE6. Site A is located in a cleft formed in between two neighboring E1-E2 heterodimers (Fig. 1a–c), a similar location where LDLRAD3 binds to VEEV25,26. Site B is located near the connecting β strands in between the A and B domains of the EEEV PE6 E2 protein (Fig. 1a–c). Site B is adjacent to site A. Densities at site B are much weaker compared with these at site A. Weak densities were observed in between sites B and A at a low contouring level (Fig. 1b). Site C is on the B domain of E2 and is approximately 60 Å and 40 Å away from sites A and B, respectively (Fig. 1a–c). The densities on the three binding sites are good enough for model fitting but not for ab initio model building of the bound VLDLR LAs (Supplementary Fig. 2b). We, thus, were not able to unambiguously distinguish the bound LAs from the 8 LA repeats. Of note, densities at different A sites in the asymmetric unit show distinct different features (Fig. 1d).
a A surface-rendered representation showing the density map of EEEV PE6 VLP bound with LA1-8-Fc. The E1, E2, and capsid of the EEEV PE6 VLP are colored coral, wheat, and thistle, respectively. Additional densities at two different locations on the spike, which could be contributed by the bound LA repeats, are colored blue and green, respectively. The white triangle indicated one asymmetric unit of the E protein shell. The outlines of four E1-E2 heterodimers in one asymmetric unit are shown in yellow. The labels i3 and q3 indicate one i3 spike and one q3 spike, respectively. b A surface-rendered representation showing the density map of one asymmetric unit. The density map was obtained from the block-based reconstruction that treats one asymmetric unit as an individual block. The E1 and E2 glycoproteins are colored the same as in “a”. The additional densities at three different sites on the spike are colored blue, green, and magenta, respectively. The zoom-in inserts show the densities at sites B. The density map in the zoom-in inserts was low pass filtered to a resolution of 10 Å and the contouring level was set to 0.0035 e−/Å3. c A cut-off view of the spike showing the positions of E1 and E2 glycoproteins, capsid, and the bound LA repeat at sites A, B, and C. The density map is set transparent. The fitted structures are shown in ribbon with E1, E2, and capsid proteins colored the same as in “a”. Fitting of the LA3 structure (pdb accession number: 8IHP) into the cryo-EM density showing the orientation and position of the bound LA repeats, which are colored blue, magenta and green, respectively. d Comparison of the LA densities at the four A sites. LA densities at the four A sites are colored gray, cornflower blue, blue, and cyan, respectively. e Ribbon diagrams show the interactions between the E protein and the bound LAs. The bound LAs at site A1, A3, B3, and C2 are colored gray, blue, magenta, and green, respectively. The potential key residues involved in the interactions are shown in sticks. The calcium ions of the bound LAs are shown as green balls.
Structure predictions of the LAs27,32 showed that they have a similar structure with an elliptic cylinder shape (Supplementary Fig. 2c). The LA has two ridges and two flat surfaces (Supplementary Fig. 2d). Differences between the LAs are mainly located at the loops in between the β strands and the N- and C-termini (Supplementary Fig. 2e). One of the flat surfaces contains the calcium-binding site that is rich in negatively charged residues (NC surface). By fitting the structure of LA327 into the additional densities at the three sites, we could establish the orientation and contact interface of the bound LAs. The bound LAs at the four A sites in the asymmetric unit have two major different orientations, of which one has a rotation of ~56° relative to the other around the calcium-binding site (Fig. 1a–d). The two different orientations of the bound LAs suggests that the binding of probably different LAs at different A sites. In both orientations, the bound LAs have the NC surface buried in the cleft. Residue K156 of E2 is in close proximity to the negatively charged residues around the calcium-binding site of the fitted LAs and could be the key residue in contact at site A (Fig. 1e). Fitting at site B showed that the bound LA is in contact with the β-ribbon connector and the N-terminal helix in the domain A of E2 (Fig. 1b, c, e). Similar to the LA at site A, the NC surface is buried at the interface. Residue K231 of the β-ribbon connector is in close proximity to the calcium-binding site and could be the key residue that mediates the interactions. Given that connecting densities between sites A and B can be observed, the bound LAs on sites A and B could be two consecutive LA repeats. Fitting at site C showed that the bound LA has close contacts with the E2 B domain, again with the NC surface buried at the interface (Fig. 1e). Similarly, a positively charged residue, K206 of E2, is in close proximity to the negatively charged residues around the calcium binding site and could be the key residue that mediates the interactions (Fig. 1e).
Mapping of the essential LAs for binding EEEV PE6
To investigate the binding of each single LA to EEEV PE6, we coated single LAs on the probe and measured their direct interactions with the EEEV PE6 VLP by using biolayer interferometry (BLI). BLI is highly sensitive to the thickness change upon the attachment of the ligand33. The binding of EEEV VLP on the probe produces significant signals due to its big size. In addition, the multivalent interactions between the dense LA molecules on the probe and the EEEV VLP allow the detection of weak binding by a single LA (Supplementary Fig. 3a). Thus, this method can be used for qualitative detection of weak interactions between each single LA and EEEV VLP17. The BLI results showed that LA1-Fc, LA2-Fc, LA3-Fc, LA5-Fc, and LA6-Fc can bind the EEEV PE6 VLP, whereas LA4-Fc, LA7-Fc, and LA8-Fc do not have detectable signal of the bound EEEV PE6 VLP (Supplementary Fig. 3a).
Based on the analyzes with the fitted LAs, we further mutated the potential key residues on the VLP and produced the mutant EEEV PE6-K156A, EEEV PE6-K206A, and EEEV PE6-K231A VLPs, which are supposed to disrupt the binding at sites A, C and B, respectively. These mutant VLPs were then used for detecting site-specific interactions with the LAs by BLI. The results showed that LA1-Fc and LA5-Fc can not bind the EEEV PE6-K156A VLP (Supplementary Fig. 3a), in which the binding site A in the cleft has been potentially disrupted. LA1-Fc, LA2-Fc, LA3-Fc, LA5-Fc, and LA6-Fc can still bind the EEEV PE6-K206A and EEEV PE6-K231A VLPs, in which either the binding site C or B has been potentially disrupted (Supplementary Fig. 3a). Additionally, we produced the E2 domain B and detected the interactions of E2 domain B with individual single LAs by using BLI. The results showed that none of the single LAs can bind the E2 domain B with a detectable signal, indicating that single LA may only have undetectable weak interactions with site C on the E2 domain B (Supplementary Fig. 3a). LA1-8-Fc binds the E2 domain B with a KD of only 1.4 μM (Supplementary Fig. 3a), suggesting that multiple consecutive LAs can establish weak interactions with the E2 domain B or site C.
To further investigate the site preference of LA2, LA3, and LA6, we produced the mutant EEEV PE6-K156A-K231A, EEEV PE6-K206A-K231A, and EEEV PE6-K156A-K206A VLPs, of which only one functional binding site was kept. BLI analyzes with the VLPs carrying the double mutations showed that none of the LAs binds EEEV PE6-K156A-K206A (Supplementary Fig. 3a). Since EEEV PE6-K156A-K206A has, presumably, only site B for binding the receptor, indicating that binding to the site B may require the synergistic binding of the neighboring site A. In addition, the results also imply that for single LAs, no additional detectable binding site exists except for the three sites observed in the reconstruction. Furthermore, the results from EEEV PE6-K156A-K206A confirmed the essential role of K156 and K206 at site A and site C, respectively. The results with EEEV PE6-K156A-K231A are similar to these obtained with EEEV PE6-K156A (Supplementary Fig. 3a). LA1, LA2, LA3, LA5 and LA6 all can bind EEEV PE6-K206A-K231A (Supplementary Fig. 3a), of which only site A is functional. These data combined indicate that LA1 and LA5 specifically bind site A, whereas LA2, LA3, and LA6 can bind both sites A and C and binding of site B probably requires more than one LA (Fig. 2a).
a A schematic diagram showing the ectodomains of VLDLR. The LA repeats are colored according to their binding specificity to the three sites on the spike of EEEV PE6. b BLI analysis showing the binding of VLDLR fragments that contains either 2 consecutive LAs (2LAs) or three consecutive LAs (3LAs) to the EEEV PE6 VLP. Data from one representative measurement of two independent experiments are presented. For each sample, the KD was calculated by averaging the values from the two independent experiments. Source data are provided as a Source Data file.
Interactions between consecutive LA repeats and EEEV PE6
The BLI data with single LAs suggested that the binding at site B may involve more than one LA, most likely two consecutive LAs that occupy both sites A and B. We further investigated the interactions between consecutive LA repeats and the EEEV PE6 VLP by using BLI. Constructs that contain LA7 or LA8 were not made since these LA repeats showed no detectable interaction with the EEEV PE6 VLP (Supplementary Fig. 3a) and are located at the membrane-proximate end of the receptor. To measure the binding kinetic parameters precisely, we coated the anti-EEEV-E2 antibody EEEV-14314 on the probe. Then the VLPs were coated on the probe through the antibody and used for detecting the interactions with the LA repeats. By using this method, the binding of LA1-8 to the EEEV PE6 VLP was measured and the results indicate a strong binding with a KD of 2.9 nM (Fig. 2b).
We made constructs that have two consecutive LAs (2LAs) and then measured their interactions with the EEEV PE6 VLP. The results showed that the 2LAs, including LA1-2-Fc (KD: 505.4 nM), LA2-3-Fc (KD: 1.8 μM), LA3-4-Fc (KD: 3.9 μM), LA4-5-Fc (KD: 11.9 μM) and LA5-6-Fc (KD: 4.2 μM), have only weak interactions with the EEEV PE6 VLP (Fig. 2b). Among these, LA1-2-Fc has a relatively higher binding affinity.
Site C is far away from either site A or site B. At least three or four consecutive LAs would be required for simultaneous occupy of sites A and C or sites B and C. We next measured the interactions between fragments that have three consecutive LAs (3LAs) and the EEEV PE6 VLP. The results showed that LA3-5-Fc can bind the VLP much stronger compared with other 3LAs, including LA1-3-Fc, LA2-4-Fc, and LA4-6-Fc. The measured KD of LA3-5-Fc is 32.3 nM, which is one to two orders of magnitude smaller than these of LA1-3-Fc (KD: 363.3 nM), LA2-4-Fc (KD: 1.1 μM) and LA4-6-Fc (KD: 2.3 μM) (Fig. 2b). The dramatic increase in the binding of LA3-5-Fc, but not other 3LA constructs, suggest possible synergistic binding through the LAs of LA3-5. The addition of a single LA repeat around LA3-5 did not further improve the binding affinity, as indicated by the measured KD values of LA3-6-Fc (KD: 71.5 nM) and LA2-5-Fc (KD: 42.5 nM) (Supplementary Fig. 3b). However, further addition of the LA repeat at either the N-terminus of LA2-5 (LA1-5-Fc) or the C-terminus of LA2-5 (LA2-6-Fc) improved the binding affinity to a level comparable to that of LA1-8-Fc (Supplementary Fig. 3b), indicating that compared with LA3-5, the introduction of two more additional LAs may create new binding modes. We also measured the binding of ApoER2iso2 LA1-3-Fc to the EEEV PE6 VLP and the results showed only weak binding with a KD of 704.8 nM (Supplementary Fig. 3b).
Structures of the VLDLR fragments in complex with EEEV PE6 VLP
The higher binding affinities of LA1-2-Fc and LA3-5-Fc suggest that these fragments may mediate different binding modes of the receptor. To further investigate the detailed binding modes of the LA repeats to EEEV PE6, we selected LA1-2-Fc and LA3-5-Fc for cryo-EM structural studies (Supplementary Figs. 4, 5, Supplementary Table 2).
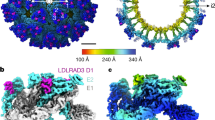
The reconstruction of LA1-2-Fc shows strong additional densities on sites A and B (Fig. 3a). By treating the E1 and E2 glycoproteins in one asymmetric unit of the icosahedral shell as a block, the reconstruction of the LA1-2-Fc and EEEV PE6 VLP complex was calculated to a resolution of 3.4 Å (Supplementary Fig. 4). Based on the density map, we identified LA1 at site A and LA2 at site B (Supplementary Fig. 4e–g). The structure model of LA1 could be built based on the density map (Supplementary Fig. 4e). Densities for the bound LA2 at site B are relatively weaker compared with these at site A. However, the key residues at the contact interface could be clearly identified (Supplementary Fig. 4f). LA1 is partially buried in the cleft created by two neighboring E1-E2 heterodimers, and the long axis of LA1 is oriented approximately 30 degrees away from the 3-fold axis of the spike (Fig. 3a-c). LA1 interacts with the E1 and E2 glycoproteins through mainly the two ridges (Fig. 3c). The negatively charged surface of LA1, which involves the calcium-binding site and is located near one ridge, is buried and have direct contacts with the positively charged residue K156 of E2 (Fig. 3d, f). Disruption of the interaction between D55 in the calcium-binding site and K156 of E2 completely abolished the binding of LA1-2 (Supplementary Fig. 3b). W50, which is in proximity to the calcium-binding site of LA1, packs in parallel against the key residue E2-K156 (Fig. 3d). Previous studies showed that the tryptophan residue in proximity to the calcium binding site is critical for the binding of the LA repeat to SFV and VEEV25,27. To verify the role of W50, we made the mutant LA1-2-W50A-Fc and BLI analysis showed that W50A completely abolished the binding of LA1-2-Fc to the EEEV PE6 VLP (Fig. 2b). Residues K32, T41-R44 at the other ridge of LA1 have direct interactions with the fusion loop of E1 and residue H175 of E2, constituting the other major contact interface with the EEEV PE6 VLP (Fig. 3e). LA2 at the site B has only a small contact surface area of approximately 451 Å2 with the E2 protein (Fig. 3g). The calcium-binding site of LA2 is buried at the interface. K231 of E2 is pointing to the center of the calcium-binding site, where D92, D94, and D96 of LA2 are in close proximity to K231 (Fig. 3g). To further verify the critical role of residues E2-K156 and E2-K231, we measured the binding of LA1-2-Fc to PE6-K156A and PE6-K231A. The BLI results showed that both mutations significantly affected the binding of LA1-2-Fc (Supplementary Fig. 3b).
a A surface-rendered representation showing the density map of the EEEV PE6 VLP bound with LA1-2-Fc. The E1, E2, and capsid of the EEEV PE6 VLP are colored the same as in Fig. 1. Additional densities at three different locations on the spike, which could be contributed by the bound LA repeats, are colored blue, magenta, and green, respectively. The labels i3 and q3 indicate one i3 spike and one q3 spike, respectively. b A surface-rendered representation showing the density map of four E1-E2 heterodimers in one asymmetric unit. The density map was calculated with the block-based reconstruction. The E1 and E2 glycoproteins and the additional densities at three different sites on the spike are colored the same as in (a). c The bound VLDLR LA1-2 on the E1 and E2 glycoproteins of the EEEV PE6 VLP. The structures of VLDLR LA1-2 are shown in the ribbon, and the structures of E1 and E2 glycoproteins are shown on the surface. The adjacent E1 protomer, E2 protomer, and the fusion loop are colored deep pink, pink, and teal, respectively. The density map of VLDLR LA1-2, which is set transparent, was low pass filtered to a resolution of 6 Å and the contouring level was set to 0.00816 e−/ Å3. d, e Detailed contacts between VLDLR LA1 and the EEEV PE6 E1 and E2 glycoproteins at site A. f Ribbon and surface-rendered representations showing contacts between VLDLR LA1-2 and the EEEV PE6 E1 and E2 glycoproteins. The surface of the E1 and E2 glycoproteins is colored according to the surface electrostatic potential. g Ribbon and surface-rendered representations showing contacts between the VLDLR LA2 and the EEEV PE6 E2 at site B. The surface of LA2 is colored according to the surface electrostatic potential.
The reconstruction of the LA3-5-Fc and EEEV PE6 VLP complex was calculated to a resolution of 3.4 Å (Supplementary Fig. 5). The result shows additional densities only on sites A and C, indicating that the binding mode of LA3-5-Fc is distinctly different from that of LA1-2-Fc (Fig. 4). By fitting the structures of LA3 and LA5 into the additional densities, we could identify LA5 rather than LA3 at site A, based on the densities around residues F198 of LA5 and I118 of LA3, and V211 of LA5 and R133 of LA3 (Supplementary Fig. 5e), which are the residues at the equivalent positions of LA3 and LA5, respectively, but have distinct different features (Supplementary Fig. 5e). The results are consistent with the BLI analysis, which showed that LA5 is site A specific (Supplementary Fig. 3a). The atomic model of the bound LA5 could be built based on the density map (Supplementary Fig. 5f). LA5 binds site A mainly through only one ridge and the buried surface area at the interface is only 302 A2. The negatively charged residues around the bound calcium of LA5 are buried and have close contacts with residue K156 of E2 (Fig. 4d). Although the major area that LA1 and LA5 attach to is similar, LA5 binds site A in a distinctly different orientation when compared with that of LA1. The long axis of the bound LA5 is almost parallel to the axis of the trimeric spike (Fig. 4e). The structure of LA3 could be well fitted into the additional densities at site C and densities of the residues at the contact interface could be clearly observed (Supplementary Fig. 5g). At the contact interface, residues Q126, D135, E137 and D139 of LA3 form salt bridges and hydrogen bonds with residues of the E1 and E2 glycoproteins, while W132 of LA3 is parallelly packed with residue K206 and V204 of E2 (Fig. 4f). Compared with other LAs, residue E137 of LA3 is unique (Supplementary Fig. 6a) and could help to establish additional salt bridges with K61 of E1 and K206 of E2 (Fig. 4f). However, mutating E137 to the residue D137 as in other LAs did not affect the binding affinity of LA3-5-Fc (Supplementary Fig. 3b). At a low contouring level, densities for LA4 could be observed in between the LA3 on one spike and the LA5 on a neighboring spike (Fig. 4c), indicating that LA3-5 crosslinks two neighboring spikes. Further mutagenesis studies showed that W132G of the LA3 abolishes the binding of LA3-5 to the EEEV PE6 VLP (Fig. 4g), indicating that W132 is the key residue of LA3 for binding EEEV PE6. Diminishing the hydrogen bond mediated by residue D135 of the LA3, D213, or D217 of the LA5 significantly reduced the binding affinity of LA3-5-Fc, indicating the essential role of the charge-charge interaction in binding EEEV PE6 (Fig. 4g and Supplementary Fig. 3b). These results also confirm that binding of LA3-5 to EEEV PE6 requires the synergistic action of both LA3 and LA5. The binding of LA3-5 requires the A site as does LA1-2, indicating that these fragments can compete with each other for binding site A. To further verify the critic role of residue E2-K206, we measured the binding of LA3-5-Fc to PE6-K206A. The BLI results showed that the mutation K206A significantly affected the binding of LA3-5-Fc (Supplementary Fig. 3b).
a A surface-rendered representation showing the overall structure of the complex. The E1, E2, and capsid of EEEV PE6 VLP are colored the same as in Fig. 1. Additional densities at two different locations on the sites A and C of the spike, which could be contributed by the bound LA repeats, are colored cyan and green, respectively. The labels i3 and q3 indicate one i3 spike and one q3 spike, respectively. b Ribbon and surface-rendered representations showing the density map of one asymmetric unit. The density map was obtained from the block-based reconstruction that treats one asymmetric unit as an individual block. The E1 and E2 glycoproteins and the additional densities at two different sites on the spike are colored the same as in (a). c Ribbon and surface-rendered representations showing the bound LAs on the spike. Additional densities beyond the spike, which were set transparent, were low pass filtered to a resolution of 8 Å and the contouring level was set to 0.0025 e−/ Å3. A model of LA4 predicted by AlphaFold was fitted in the densities with the N- and C-termini of LA4 posed toward the C-termini of LA3 at site C and N-termini of LA5 at site A, respectively. d Detailed contacts between VLDLR LA5 and the EEEV PE6 E1 and E2 glycoproteins at site A. The adjacent E2 protomer is colored pink. e Structural comparisons of the bound LA1 and LA5 at site A. The bound LA1 and LA5 are colored blue and cyan, respectively. f Ribbon and surface-rendered representations showing contacts between VLDLR LA3 and the E1 and E2 glycoproteins at site C. The surface of LA3 is colored according to the electrostatic potential. The fusion loop is colored teal. g BLI analysis showing the binding of VLDLR LA3-5 mutants to the EEEV PE6 VLP. For each sample, the KD was calculated by averaging the values from the two independent experiments. Source data are provided as a Source Data file.
The multiple different binding modes of VLDLR
The combination of multiple bounders and binding sites can generate many different complex modes. The fragments LA3-5 and LA1-2 have much higher binding affinities compared with these with the same number of LA repeats. Additionally, the cryo-EM reconstructions with LA1-2-Fc and LA3-5-Fc showed that the densities of LA3-5 and LA1-2 share similar features to these observed in the reconstruction of the LA1-8-Fc-PE6-VLP complex. The fragments LA1-2 and LA3-5 could play a major role in the binding of VLDLR to EEEV PE6. With the high-resolution structures of LA1-2 and LA3-5, we further analyzed the reconstruction of the LA1-8-Fc and EEEV PE6 VLP complex, which shows strong densities at sites A and C, and weak densities at only sites B of the q3 spike. Among the three B sites on the q3 spike, the one (B3, Fig. 1a, b, d) near the 5 fold vertex has the strongest densities. The orientation of the densities at the nearby site A3 are consistent with these of the bound LA1 (Fig. 1a, b, d). In contrast, the orientation of the densities at the site A1 of the i3 spike are consistent with these of the bound LA5. Densities at site A2 and A4 are probably the average of the bound LAs. The fragment LA3-5 mediated binding is likely dominant, since the densities at sites B are much weaker compared to these on sites A and C.
It should be noted that the binding affinity of LA1-2 or LA3-5 is significantly weaker than that of LA1-8 or LA1-6, implying that, except for the LA1-2 and LA3-5 mediated binding modes, additional binding mode must exist when more than 4 consecutive LA repeats present. Sites C around the 2-fold and 5-fold axes may accommodate more than three consecutive LA repeats. To investigate possible additional binding modes with VLDLR fragments of more than 4 consecutive LA repeats, we incubated LA1-8-Fc and the EEEV PE6-K156A VLP, with which both the LA1-2 and LA3-5 mediated binding should be disabled. As expected, the cryo-EM reconstruction of the complex showed additional densities only on sites C. Further structural analysis by fitting the structure of LA3 into the density map showed that the fragments of more than three consecutive LA repeats could bind the C sites around the 2-fold and 5-fold axes (Supplementary Fig. 6b).
VLDLR mediated cell attachment of EEEV PE6
To further investigate whether the LA1-2 or LA3-5 mediated binding mode could establish stable attachment of the virus to the cell, we mutated the receptor and produced VLDLR-W132A and VLDLR-W50A, in which the key residue W132 of LA3 and W50 of LA1 were mutated to abolish the binding of LA3 and LA1, respectively. As a consequence, the LA3-5 or the LA1-2 mediated binding mode of VLDLR would be presumably disrupted. We built HEK293T cell lines that stably express either the wild-type VLDLR, the LA3-dysfunctional (W132A), or the LA1-dysfunctional (W50A) VLDLR mutant. Cell attachment assays were then performed with the stable cell lines and the AF647 labeled EEEV PE6 VLP. As expected, the efficiency of the mutant VLDLR-W50A mediated cell attachment of the EEEV PE6 VLP was significantly reduced when compared with that of the wild-type VLDLR mediated cell attachment (Fig. 5a–c). However, the efficiency of the mutant VLDLR-W132A-mediated cell attachment was significantly enhanced when compared with that of the wild-type VLDLR mediated cell attachment (Fig. 5a–c).
a Representative confocal microscopy images showing the attachment of EEEV to the HEK293T cell lines that express the wildtype VLDLR, VLDLR-W50A, and VLDLR-W132A, respectively. The EEEV-PE6 and EEEV PE6-K206E VLPs were used. SFV VLPs were used as a control. Cells were incubated with the AF647 labeled VLPs at 37 °C for 15 min and then were imaged by using a confocal microscopy. Fluorescence from the WGA-AF488 labeled (green) cell membranes and the AF647 labeled (pink) VLPs were excited by laser beams with a wavelength of 488.6 nm and the wavelength of 639.8 nm, respectively. The power and PMT gain (HV) of the laser beam at the wavelength of 488.6 nm were set to 0.6 and 40 V, respectively. The power and PMT gain (HV) of the laser beam at the wavelength of 639.8 nm were set to 2 and 88 V, respectively. The scale bar is 20 μm. b Relative attachment rate of the VLPs. n = 3 biological replicates. Bars represent relative attachment (%) ± SEM calculated with three groups of images from independent assays. Five images are in each group. Black diamond-dots indicate individual data point for biological replicates. Statistical significance was calculated by using one-way ANOVA with Tukey’s multiple comparisons test, ****P < 0.0001. Source data are provided as a Source Data file. c Western blot analysis showing the expression level of the wildtype VLDLR, VLDLR-W50A, and VLDLR-W132A in the same number of cells. Samples were run on the same blot. Uncropped and unprocessed scans of blots are provided in the Source Data file. d BLI analysis showing the binding of VLDLR LA1-8-Fc to the EEEV PE6-K206E VLP. Data from one representative measurement of two independent experiments are presented. The KD was calculated by averaging the values from the two independent experiments. Source data are provided as a Source Data file.
We then checked whether LA1-2 or LA3-5 alone can mediate the internalization of the virus. We generated two VLDLR mutant constructs: LA1-8 (LA1-2)-Fc and LA3-8 (MutLA6)-Fc. In the construct of LA1-8(LA1-2)-Fc, only LA1 and LA2 (the two LAs in the parenthesis) retain the key residues W50 and W89 unmutated, while all corresponding residues in the other LAs were mutated to alanine. Therefore, this mutant can be used to verify the function of LA1-2. In the construct of LA3-8(MutLA6)-Fc, LA1-2 was removed and LA6 was mutated to loss the binding ability. Thus, only LA3-LA5 are functional in recognizing EEEV-VLP. Consequently, this mutant can be used to verify the function of LA3-5. Next, we overexpressed the two VLDLR mutants in HEK293T cells and subsequently incubated the cells with AF647-labeled VLPs. The cells were then scanned by using a live cell confocal microscopy (ZEISS 980) at the indicated temperatures. The results showed that both mutants can mediate the attachment and internalization of EEEV (Supplementary Fig. 7). Thus, we conclude that both the LA1-2 and LA3-5 mediated binding modes enable cell surface attachment and internalization of EEEV.
To explore the mechanism of the VLDLR-W132A mediated enhancement of cell attachment, we analyzed whether the mutant W132A affects the binding of VLDLR to the receptor-associated protein (RAP), a ligand of the LDL-related receptors known to inhibit the binding of EEEV, SFV, and SINV17. We used the Fc tag of VLDLR to pull down RAP from cells that overexpress RAP and the indicated VLDLR. Consistent with a previous report34, we observed a significantly weaker band of RAP bound to the mutant VLDLR-W132A-Fc compared to that bound to the wild type VLDLR-Fc (Supplementary Fig. 6d). The diminished binding to RAP could enhance the binding of VLDLR-W132A to viruses. As shown in the results of the cell attachment assays, the efficiency of the mutant VLDLR-W132A mediated cell attachment was enhanced for both the EEEV VLP and the SFV VLP (Fig. 5). However, the enhancement observed with the EEEV PE6 VLP is significantly greater than that with the SFV VLP, suggesting the existence of additional mechanisms for EEEV.
We further explored other possible mechanisms involved in the VLDLR-W132A-mediated enhancement of EEEV cell attachment. We produced the mutant LA1-6-W132A-Fc. BLI analysis indicated that the mutation W132A does not affect the binding of LA1-6 (LA1-6-W132A-Fc, KD: 2.6 nM) (Supplementary Fig. 3b), suggesting that the loss of function in LA3 could be compensated by other LAs. Cryo-EM reconstructions of the LA1-6-Fc-W132A and EEEV PE6 VLP complex were then calculated (Fig. 6). The results show strong additional densities only on sites A and B and the densities on sites A and B are equally strong. The density distribution is similar to that of the LA1-2-Fc-PE6-VLP complex, indicating that the LA1-2 mediated binding to EEEV PE6 is dominant with the mutant LA1-6-Fc-W132A. The introduction of the dysfunctional LA3 in VLDLR drives the switch from the LA3-5-dominated binding mode to the LA1-2-dominated binding mode. LA3 is the determinant of the binding mode and may interfere with the binding of LA1-2. The conclusion could be further supported by the mutant LA1-3-W132A-Fc (KD: 279.0 nM) (Supplementary Fig. 3b), which has slightly higher binding affinity to the EEEV PE6 VLP compared with that of the wild-type fragment LA1-3-Fc (KD: 363.3 nM) (Fig. 2b). Furthermore, we determined the cryo-EM structure of LA1-3-Fc in complex with the EEEV PE6 VLP and checked the possible disturbance on the binding of LA1-2 by LA3. Compared with the reconstruction of the LA1-2-Fc and EEEV PE6 VLP complex, the densities at sites B were largely eliminated in the reconstruction of the LA1-3-Fc and EEEV PE6 VLP complex (Fig. 6), indicating that LA3 can compete with LA1 for binding site A and disturbs the binding of LA1-2. Similarly, we did the cryo-EM structural studies of the LA2-3-Fc-PE6-VLP complex (Fig. 6). We observed additional densities only at sites A. Comparison of the density maps showed that the bound LA has a similar orientation as that of LA5 (Supplementary Fig. 6c). Given that both LA2 and LA3 can bind site A, the densities at sites A could be the average of LA2 and LA3. No matter LA2 or LA3 binds to site A, the other LA is highly flexible and not visible, which is quite different when compared with the binding of LA1-2.
Surface-rendered representations showing the overall structures and the asymmetric unit of the EEEV PE6 VLP in complex with VLDLR LA1-8-Fc, LA1-2-Fc, LA3-5-Fc, LA1-3-Fc, LA2-3-Fc or LA1-6-W132A-Fc, and the EEEV PE6-K206E VLP in complex with LA 1-8-Fc. The densities of E1 and E2 glycoproteins are colored dark gray and light gray, respectively. The additional densities at sites A, B and C are colored blue, magenta and green, respectively. The density maps of the asymmetric unit were low passed to 6 Å and normalized with the densities of the selected region of E2 (residues 80-120), which are located at the central region of the spike.
Taking together, these data combined indicate that LA3 can compete with LA1 for binding site A and thus, disturbs the LA1-2 mediated stable interactions with the virion. Both the LA1-2 and the LA3-5 mediated interactions could establish stable attachment of the virion to the cell, but the LA1-2 mediated attachment may mediate virus attachment more efficiently, due to probably the defined orientation of the bound receptor created by LA1-2. The C sites around the 5-fold and 2-fold symmetry axes generate additional binding modes of VLDLR, which could also play an important role in mediating the virus entry.
Binding of VLDLR to different EEEV strains
Of note, sequence alignments indicate that the envelope proteins of the PE6 strain have two unique mutations when compared with these of other EEEV strains, including the North and South American strains (Supplementary Figs. 8, 9). One of the unique mutations is the residue K206 of E2, which is a glutamic acid residue (E) in other virulent strains. K206 is the key residue of site C, while the residue E206 may abolish or significantly weaken the binding of VLDLR at site C. We, thus, mutated K206 to E206 and made the mutant EEEV PE6-K206E VLP. BLI analysis showed that the binding of LA1-8-Fc was significantly affected by the mutation K206E. LA1-8-Fc binds to the EEEV PE6-K206E VLP with a KD of 167.0 nM (Fig. 5d). In contrast, the KD with the EEEV PE6 VLP is 2.9 nM (Fig. 2b). Further cryo-EM structural analysis of LA1-8-Fc in complex with the EEEV PE6-K206E VLP showed that similar additional densities on sites A and B as these of the LA1-8-Fc-PE6-VLP complex (Fig. 6). However, no additional densities could be observed on sites C, indicating that the C sites are impaired by the mutation K206E.
We next measured the VLDLR-mediated cell attachment with the EEEV PE6-K206E VLP. The results showed that the mutation K206E significantly reduced the cell attachment of EEEV (Fig. 5a–c and Supplementary Fig. 10), which is in consistent with the BLI analysis (Figs. 2b, 5d). We further measured the VLDLR-W132A mediated cell attachment of the EEEV PE6-K206E VLP. The results showed that the mutation W132A promoted approximately 40 times more VLPs attached to the cells when compared with that of the wild type VLDLR mediated cell attachment (Fig. 5a–c and Supplementary Fig. 10).
Discussion
Sequence alignments show that the key residues involved in binding VLDLR at sites A and C are highly conserved among LA1, LA2, LA3, LA5, and LA6 (Supplementary Fig. 6a). Comparing to the LAs that can bind site C, LA1 and LA5 have different residues at positions 44 and 47 of LA1 and positions 204 and 214 of LA5. Mutagenesis studies showed that LA1-R44Q-T47P-Fc and LA5-S204Q-D214G-Fc can bind EEEV PE6-K156A (Supplementary Fig. 11a), indicating these are the key residues that prevent the binding of LA1 and LA5 at site C. Structural superimpositions showed that LA2 and LA3 could bind site A in a similar way as that of LA5 (Supplementary Fig. 6c).
The bound LA1 places its C terminus towards site B, facilitates the binding of LA2 at site B, and promotes specific strong binding of LA1-2 at sites A and B, which distinguishes LA1-2 from other 2LAs (Figs. 2b, 3c). In addition, LA1-2 defines a unique orientation of the receptor that could favor the binding of cofactors on the cell membrane and help to build the stable attachment of the virus.
Site A and site C are 60 Å away from each other and they can only be simultaneously occupied by two LAs of a VLDLR fragment that contains at least three LAs. The location of the N and C-termini of the bound LAs further confines the binding of an N-terminal LA at site A from one spike and the binding of a C-terminal LA at site C from a neighboring spike. Among the 3LAs, only LA3-5 can satisfy the requirements for binding sites A and C at the same time, since LA1 is site A specific, while LA4 can not bind either site A or site C.
In conclusion, the difference in the binding affinity and specificities of each LA and the geometric restrains created by the arrangement of the viral glycoproteins, define the fragment-mediated distinct binding modes of VLDLR to EEEV. The EEEV PE6 strain was isolated from the brain of an infected horse and was propagated in chicken embryos35,36. The E206K mutation of E2 in EEEV PE6 was considered as the key factor of attenuation since domain B was identified as the epitope of neutralization antibodies31. However, our results showed that the E206K mutation creates an additional binding site of the receptor and significantly enhances the VLDLR-mediated cell attachment of EEEV (Fig. 5a–c, Supplementary Fig. 6b). The other unique mutation identified in the EEEV PE6 envelop protein, F311 of E1, is not relevant to the binding of VLDLR (Supplementary Fig. 8). The key mutation that causes the attenuation of EEEV PE6 could be in the none structural proteins rather than the structural proteins.
There are advantages and disadvantages of using the EEEV PE6 strain as vaccines. In its inactivated or VLP form, EEEV PE6 strain could provide protection against the additional C site at the domain B of E2, which could exist in the circulating enzootic EEEV strains. However, if being used as the live attenuated vaccine, EEEV PE6 strain can have the risk of being converted into a strain of strong virulence, due to its high efficiency in binding VLDLR.
Of note, we observed that the VLDLR mediated cell attachment of EEEV was significantly enhanced with the mutation W132A, which results in a dysfunctional LA3. The enhancement was observed for both the EEEV PE6 and PE6-K206E VLPs (Fig. 5a-c, Supplementary Fig. 10) and the promotion on PE6-K206E is dramatical. Previous studies also showed that the absence of the LA3 repeat could confer beneficial functional changes to VLDLR37. A naturally occurring VLDLR variant (VLDLR-III) lacking the LA3 repeat is more effective in lipoprotein binding in vitro and lipoprotein uptake in vivo compared to that of the full length VLDLR37.
Considering that E206 is dominant in the current virulent EEEV strains, our finding might be of physiological importance. The mutation W132G, which is supposed to behave similarly as W132A, can be identified in the human genome and SNP sequences. W132G was shown to abolish the binding of LA3-5-Fc to the EEEV PE6 VLP (Fig. 4g). We further made the mutant VLDLR-W132G and the cell attachment assays showed similar results as these with the mutant VLDLR-W132A (Supplementary Fig. 11b). It would be interesting to investigate in the future whether human carrying the W132G mutation in VLDLR or other similar mutations in LA3 are more susceptible and of higher risk to EEEV infection.
Previous studies showed that the VLDLR from S. vulgaris, the reservoir host of EEEV, can not mediated the infection of K562 cells by EEEV17. However, sequence alignments of the VLDLRs from different species showed that key residues of the S. vulgaris VLDLR are exactly the same as these of the human VLDLR (Supplementary Fig. 12). We further measured the binding of S. vulgaris VLDLR LA1-8-Fc (LA1-8-Fc-S. vulgaris) to the EEEV PE6 VLP and the results showed that LA1-8-Fc-S. vulgaris binds the EEEV PE6 VLP with a KD of 11.0 nM, which is similar as that of the human LA1-8-Fc (Supplementary Fig. 13a and Fig. 2b).
We then investigated the localization of the overexpressed S. vulgaris VLDLR in HEK293T cells and the results showed that the distribution of the receptor has no significant difference when compared with that of the human VLDLR (Supplementary Fig. 13b). Next, we checked the attachment of the EEEV PE6 VLP to the HEK293T cells that overexpress the S. vulgaris VLDLR and the results showed that S. vulgaris VLDLR can mediate the stable attachment of the EEEV PE6 VLP. Furthermore, the VLDLR mediated entry of the EEEV PE6 VLP was checked and the results showed the labeled EEEV PE6 VLP particles in the cell (Supplementary Fig. 13c). The binding of EEEV PE6-K206E to the S. vulgaris VLDLR+ HEK293T cells is weak but can be detected (Supplementary Fig. 13d). These results indicate that the S. vulgaris VLDLR is able to mediate the attachment and entry of EEEV. However, different EEEV strains may have different susceptibility to the bird hosts.
Sequence alignments of VLDLRs from the vertebrate species show that the key residues involved in the interactions with sites A, B and C are completely conserved in most of the LA repeats (Supplementary Fig. 12). Thus, the LA1-2 and LA3-5 mediated binding of VLDLR to EEEV could be well maintained in most vertebrate species. Larger variations are observed in the sequences of VLDLR from invertebrate species when compared with these among the vertebrate species (Supplementary Fig. 12). However, residues involved in direct contacts with EEEV are still largely conserved in the invertebrate VLDLRs (Supplementary Fig. 12), suggesting that the binding modes of VLDLR could be well maintained for mediating the infection of invertebrate vectors. The different binding modes of VLDLR involve different LAs. Loss of any functional LA may only drive the switch between different binding modes and does not affect the attachment of EEEV to the cells. Thus, EEEV could be tolerant to the variations in VLDLRs from different species. The mechanism involved in the tolerance of EEEV to the variation of VLDLR is quite different from that of SFV. LA1-5 of VLDLR binds multiple E1-DIII sites of SFV through a synergistic mode, in which the binding affinity of a single LA could be sacrificed for maximizing the overall binding affinity built on multiple sites27.
Sequence alignments of alphavirus E2 proteins show that the key residues involved in binding VLDLR is not conserved among E2 proteins of alphaviruses (Supplementary Fig. 14a). Thus, it is unlikely that VLDLR can mediate the entry of other alphaviruses in a similar way as does for EEEV.
Comparisons of the binding of the LA repeat to different viruses, the binding modes are quite different even at the similar site, such as the cleft in between the E1-E2 heterodimers. The D1 domain of LDLRAD3 binds VEEV at a deeper position of the cleft, with significantly more contacts, and much higher affinity when compared with the binding of LA1 or LA5 to EEEV (Supplementary Fig. 14b). However, a common feature is that a positively charged residue of the virus surface protein interacts with the negatively charged residues around the calcium-binding site of the LA repeat. The surface in contact could be as small as 245 Å2, such as those between LA3 and the E2 B domain of EEEV. The LA repeat could be a modular scaffold for specific binding of many different molecules, through either a single LA repeat or multiple LA repeats. In these involved multiple similar repeating subunits, although the binding affinity of each single LA is weak, simultaneous interactions at multiple sites could build up enough affinity for highly specific recognition of the target molecules, especially for these densely arranged molecules, such as viral envelop proteins and capsids.
Methods
Cell culture and treatments
HEK293T cells were obtained from ATCC (Cat# CRL-3216) and cultured at 37 °C in the Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco), 25 mM HEPES (Thermo Fisher Scientific), 1% (v/v) penicillin-streptomycin (Thermo Fisher Scientific) and 2 mM GlutaMax (Thermo Fisher Scientific) with 5% CO2. HEK293F cells were obtained from Thermo Fisher Scientific (Cat# R79007) and cultured at 37 °C in the medium SMM 293-TII (Sino Biological Inc., M293TII-1) with 5% CO2.
EEEV VLP production and purification
The genes encoding the EEEV structural proteins (PE6, GenBank accession number AAU95735.1), including capsid, E1, E2 and 6 K, were codon-optimized and synthesized (Ruibiotech). The synthesized genes, with the K67N mutation in the capsid protein as previously reported38, were cloned into the vector pCMV to produce the EEEV PE6 VLPs. EEEV VLPs were produced in HEK293F cells through transient expression of the structural proteins. Approximately 60 h after transfection, the supernatant was harvested by centrifugation at 1000 × g for 15 min and further centrifugation at 4000 × g for 30 min to remove the cell debris. The VLPs were collected at 4 °C with a 30% (w/v) sucrose cushion by centrifugation at 110,000 × g for 2.5 h in a SW32Ti rotor (Beckman). The pellet was resuspended in a buffer containing 20 mM HEPES at pH 8.0 and 150 mM NaCl. The VLPs were further purified by ultracentrifugation on a 20–60% (w/v) sucrose density gradient with a SW41 rotor (Beckman) at 140,000 × g for 15 h at 4 °C. The VLP band was collected from the gradient using a syringe. The sucrose was removed through buffer-exchange with 20 mM HEPES at pH 8.0 and 150 mM NaCl. The purity and integrity of the VLPs were examined by negative-staining electron microscopy. The plasmids for producing mutant VLPs were constructed by site-directed mutagenesis.
Expression of VLDLR fragments and protein purification
To generate recombinant VLDLR fragments, plasmids encoding the VLDLR fragments were co-transfected with a vector that expresses the human RAP as described previously17. The fragments constructed include VLDLR LA1-8 (residues 1-355, GenBank accession number NP_003374.3), LA1 (residues 1-69), LA2 (residues 70-110), LA3 (residues 111-151), LA4 (residues 152-190), LA5 (residues 191-236), LA6 (residues 237-275), LA7 (residues 276-315), LA8 (residues 316-355), LA1-2 (residues 1-113), LA2-3 (residues 70-151), LA3-4 (residues 111-190), LA4-5 (residues 152-236), LA5-6 (residues 191-275), LA1-3 (residues 1-152), LA2-4 (residues 70-190), LA3-5 (residues 111-236), LA4-6 (residues 152-275), LA1-4 (residues 1-190), LA2-5 (residues 70-231), LA3-6 (residues 111-276), LA1-5 (residues 1-236), LA2-6 (residues 70-275), and LA1-6 (residues 1-275). The insertions were fused at the C-terminus with a pre-inserted region encoding a human IgG1 Fc. A HRV 3 C protease cleavage site (GPLEVLFQGP) was introduced in between the insertion and the Fc tag. The authentic VLDLR signal sequence was used and placed at the N-terminus of the insertions. The mutations were constructed by site-directed mutagenesis. The RAP gene was cloned into the vector pCMV. HEK293F cells were used for the expression of the VLDLR fragments. Supernatant was collected 72 h after transfection by centrifugation at 4000 × g for 30 min. The recombinant proteins were purified by using the protein A Sepharose 4B resin (GE Healthcare, 17-0780-01). To remove RAP from the VLDLR fragment on the protein A resin, the resin was washed with 100 column volumes of 10 mM EDTA, 20 mM HEPES at pH 8.0, 150 mM NaCl and 50 column volumes of 10 mM EDTA, 20 mM HEPES at pH 8.0, 500 mM NaCl. After the wash, the target proteins were refolded on the column with 100 column volumes of the refolding buffer containing 2 mM CaCl2, 20 mM HEPES at pH 8.0 and 150 mM NaCl. The refolded proteins were eluted from the resin by using 0.1 M glycine at pH 2.7. The eluted protein was neutralized with 1 M Tris-HCl at pH 9.0. The elution was concentrated and further purified by size-exclusion chromatography (Superdex 200 Increase 10/300 GL, GE Healthcare). The running buffer used contains 20 mM HEPES at pH 8.0 and 150 mM NaCl.
The gene fragment encoding the EEEV PE6 E2 Domain B (E2 residues 170-232, GenBank accession number AAU95735.1) was fused at the C-terminus with an Fc for producing the recombinant E2 Domain B protein in HEK293F cells. Soluble protein was purified from the supernatant using the Protein A Sepharose 4B resins (GE Healthcare).
Pull down assays
To express VLDLR-RAP complex in HEK293F cells, plasmids encoding the VLDLR LA1-8-Fc or LA1-8-W132A-Fc were co-transfected with plasmids encoding human RAP (2:1 ratio). Supernatant was collected 72 h after transfection by centrifugation at 4000 × g for 30 min. The recombinant proteins were purified by using the protein A Sepharose 4B resin. After the wash with buffer containing 20 mM HEPES at pH 8.0 and 150 mM NaCl, the VLDLR-RAP complex was eluted from the resin by using 0.1 M glycine at pH 2.7. The eluted protein was neutralized with 1 M Tris-HCl at pH 9.0 and then analyzed using SDS-PAGE gels.
Bio-layer interferometry analysis
BLI analyzes were performed at 25 °C using an Octet Red biosensor system (Sartorius). LA1-Fc, LA2-Fc, LA3-Fc, LA4-Fc, LA5-Fc, LA6-Fc, LA7-Fc and LA8-Fc (25 μg ml−1) were loaded onto the ProA Biosensors (Sartorius 18-5010) in the loading buffer (20 mM HEPES, 150 mM NaCl, pH 8.0) for 3 min. After a baseline measurement for 180 s in the loading buffer, VLPs at a concentration of 0.1 mg ml−1 (EEEV PE6, EEEV PE6-K156A, EEEV PE6-K231A, EEEV PE6-K206A, EEEV PE6-K156A-K206A, EEEV PE6-K156A-K231A, EEEV PE6-K206A-K231A) were incubated with the probes for 300 s and the probes were then incubated in the loading buffer for 300 s.
The binding affinity of the VLDLR fragments were measured as below. Antibody EEEV-143 was mixed and incubated with biotin (EZ-Link-NHS-LC-LC-Biotin, Thermo Fisher) at a molar ratio of three biotins to one EEEV-143 at room temperature for 30 min. Excess biotin was removed using a desalting column (2-ml Zeba Spin 7 K MWCO, Thermo Fisher). Biotinylated EEEV-143 (25 μg ml−1) was immobilized onto the SA Biosensors (Sartorius #18-5019) for 3 min and then EEEV VLPs (50 μg ml−1) were incubated with the biosensor for 30 min. The tips were washed with the wash buffer (20 mM HEPES, 150 mM NaCl, pH 8.0, 0.1% BSA) for 3 min to obtain a baseline reading, then the biosensors were dipped into wells containing the VLDLR fragments at various different concentrations for 5 min, followed by a 5 min soaking in the buffer (20 mM HEPES, 150 mM NaCl, pH 8.0, 0.1% BSA) for dissociation. Data analysis was performed using the software Octet (version 12.2) with a standard 1:1 binding model and graphs presented were plotted in Prism 9 (version 9.0.0). Note that due to the use of bivalent Fc fusion constructs in the BLI assays, the calculated KD values obtained from the BLI data should be regarded as the KD apparent values.
Stable cell line generation
To generate the cell line that stably expresses the full length human VLDLR, human VLDLR mutants (VLDLR-W50A, VLDLR-W132A, VLDLR-W132G) or S.vulgaris VLDLR (GenBank accession number: XP_014736085.1), the VLDLR gene was cloned into the lentiviral vector pHAGE. HEK293T cells at a confluence of ~70% were transfected with 2 μg pHAGE, 1 μg pMD2.G, 1 μg psPAX2 and Lipofectamine 2000 (Thermo Fisher, cat# 11668019). The control cell line was transfected with the empty vector pHAGE, pMD2.G and psPAX2 (Fig. 5a, Supplementary Figs. 10, 11 and 13). The cells were cultured in a 6‐well plate with the DMEM medium (Thermo Fisher) and 10% FBS (Gibco, cat# 10091148) to generate the lentivirus. The medium containing lentiviruses was collected 60 h after the transfection. The cell debris was removed by centrifugation. The lentivirus collected was added to the HEK293T cells cultured in a 6‐well plate at a confluence of 50–60%. The virus was removed 24 h after infection and the infected cells were resuspended in fresh DMEM medium containing 10% FBS. The cells were cultured for another 24 h, and then puromycin was added to the medium at a final concentration of 2.5 μg/ml. After passaging the cells three times in medium containing 2.5 μg/ml puromycin, the cells were maintained in the medium containing 2.5 μg/ml puromycin and were used for the cell attachment assays.
Antibodies
The following antibodies were used in Western blotting. The anti-Strep-Tag II monoclonal antibody 8C12 (Cat. No. A02230, 1: 5000) were obtained from Abbkine. Antibody Pretreat Solution (HRP/Mouse) (CW2030M) were obtained from Cwbio. The anti-β actin monoclonal antibody (66009-1-IG, 1:5000) were obtained from Proteintech.
Western blotting
Cells were washed once with PBS (Gibco) and immediately lysed with the lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 × Cocktail) on ice for 5 min. Samples were collected by centrifugation at 12000 g for 5 min at 4 °C. The samples were separated with 10% polyacrylamide gels and then were transferred to the PVDF membranes (Millipore) by using a Bio-Rad Criterion transfer system. After the transfer, membranes were blocked for an hour at room temperature with the blocking buffer (5% non-fat milk in TBS-T), then were sequentially incubated for 1 hour at room temperature in the solutions that contains the diluted primary and secondary antibodies. The antibody solutions were prepared according to the manufacturer’s protocol. After three washes with TBS-T, the membranes were developed by adding the chemiluminescent HRP substrate (Bio-Rad) and were imaged by using a Chemiluminescent Imaging System. After probing, the membranes were stripped to remove the primary and secondary antibodies with a stripping buffer (NCM, Biotech) for 10 min at room temperature. After three subsequent washes in TBS-T, the membranes were used for the detection of the reference protein.
Ectopic expression of VLDLR
The human and S. vulgaris VLDLR genes were cloned into the vector pCMV that introduced a Strep-tag II (WSHPQFEK) in fusion with the C terminus of the recombinant receptor. For the localization assay, EGFP was inserted after the Strep-tag II (WSHPQFEK). To explore the LA1-2 and LA3-5 mediated binding modes, we generated two VLDLR mutant constructs based on the human VLDLR-strep-pCMV construct: LA1-8(LA1-2)-strep and LA3-8(MutLA6)-strep. In the construct of LA1-8(LA1-2)-strep, only LA1, and LA2 (the two LAs in the parenthesis) retain the key residues W50 and W89 unmutated and all corresponding residues in the other LAs were mutated to alanine (W132A, F171A, W210A and W256A). In the construct of LA3-8(MutLA6)-strep, LA1-2 was removed and LA6 was mutated to loss the binding ability (W256A). Prior to transfection, HEK293T cells were seeded in 6-well plates or in 35 mm glass bottom microwell dishes (MatTek, #P35G-1.5-14-C) at a density of 4 × 105 cells/ml. Approximately 24 h after the seeding, cells were grown to 60% confluency and were transfected with 2.5 μg plasmid using Lipofectamine 2000 (Thermo Fisher, cat# 11668019). The plasmid-Lipofectamine 2000 mixture was prepared and incubated in 250 μl Opti-MEM medium (Gibco) for at least 5 min at room temperature before being added dropwise with a pipette into the cell culture. At 24 h post-infection, cells were used for imaging.
Cell localization of S. vulgaris VLDLR
To determine the cell localization of S. vulgaris VLDLR, HEK293T cells growing in 35 mm glass bottom microwell dishes were transfected with human-VLDLR-EGFP or S. vulgaris-VLDLR-EGFP for 24 h. The cells were washed once with PBS and then imaged with a Nikon A1 HD25 confocal microscope. The probes were excited by a laser beam at a wavelength of 488.6 nm.
Cell attachment and entry assay
In order to label the VLP, the buffer of the VLP was exchanged to 0.1 M sodium bicarbonate at pH 8.3 with a pre-equilibrated ZebaTM Spin Desalting Column (Thermo Scientific #89882). The VLP was then adjusted to a concentration of 1 mg/ml for labeling. 25 μl Alexa FluorTM 647 (AF647) succinimidyl ester (NHS ester, Invitrogen #A37573) in DMSO at 1 mg/ml was added to 1 ml VLP. After an incubation for 1 hour at room temperature, the excessive reactive dye was removed through buffer exchange into PBS by using a pre-equilibrated ZebaTM Spin Desalting Column (Thermo Scientific #89882). AF647 labeled VLPs were stored on ice and used within 12 hours.
For the cell attachment assay, cells stably expressing the full length human VLDLR, human VLDLR mutants (VLDLR-W50A, VLDLR-W132A, VLDLR-W132G) or S. vulgaris VLDLR were collected by centrifugation and then resuspended in culture medium to a concentration of 5 × 105 cells/ml. 12.5 μg VLPs were mixed with 250 μl cells and incubated at 37 °C for 15 min. For the cell attachment and internalization assay, HEK293T cells that overexpress human VLDLR, S. vulgaris VLDLR, VLDLR LA1-8(LA1-2) or VLDLR LA3-8(MutLA6) were collected by centrifugation and then resuspended in culture medium to a concentration of 5 × 105 cells/ml. 25 μg VLPs were mixed with 250 μl cells and incubated at 4 °C or 37 °C for 30 min. The cell-VLP mixture was washed twice with PBS and the cell membrane was labeled with 125 μl Alexa FluorTM 488 (AF488, Invitrogen #W11261) conjugated Wheat Germ Agglutinin (WGA-AF488) at a concentration of 1 μg/ml. After an incubation of 4 mins on ice, the cells were washed twice in PBS to remove free VLP and WGA. The cells were then resuspended in 30 μl PBS and placed on ice. For imaging, the samples were placed in a 35 mm glass bottom microwell dish and images of the cells were recorded with a pixel size of 0.29 μm using a Nikon A1 HD25 confocal microscope or a ZEISS LSM 980. Fluorescence signals from the WGA-AF488 labeled cell membrane and the AF647 labeled VLP were excited by a laser beam at a wavelength of 488.6 nm and 639.8 nm, respectively. For each sample, at least five images were collected. To obtain the efficiency of the wide type VLDLR and the mutant VLDLR(W50A and W132A) mediated cell attachment, signals on the surface and within the cells were calculated using the software ImageJ (National Institutes of Health, version 1.53t), and the fluorescence signal ratio of each image was calculated with the fluorescence signal from the VLP (AF647) and the cell membrane (WGA-AF488), Relative attachment rate was determined as the follows using Microsoft Excel (version 2019MSO): Relative attachment rate (%) = (fluorescence ratio of the treatment group (wild type VLDLR, the LA3-dysfunctional (W132A) or the LA1-dysfunctional (W50A) VLDLR mutant with the labeled EEEV PE6, EEEV PE6-K206E or SFV VLP) - fluorescence ratio of the control sample)/(fluorescence ratio of the wild type VLDLR with the labeled EEEV PE6 VLP - fluorescence ratio of the control sample) × 100. Graphs were plotted in GraphPad Prism (version 7.00). All the experiments were repeated three times.
Cryo-EM sample preparation and data collection
EEEV PE6 or EEEV PE6-K206E VLPs were incubated with a VLDLR LA concatemer or a mutant VLDLR LA concatemer at room temperature for ~20 min at a ratio of three receptors per EEEV E2-E1 heterodimer. Next, a holey NiTi grid (NiTi film on Au grid, Beijing EBO Technology Limited) was used for cryo-EM sample preparation, which showed better electron conductivity than the grid with a holey carbon film39. 3 μl of the sample were applied to holey NiTi grids (300 mesh, R2/1) that had been glow-discharged in oxygen and argon for 1 min. The grids were flash cooled in liquid ethane using an FEI Vitrobot Mark IV with a blotting time of 5 s at 100% humidity and 4 °C. The data collections of the samples were performed using either a Talos Arctica microscope (Thermo Fisher Scientific) operating at 200 kV or a Titan Krios G2 (FEI) microscope operating at 300 kV. Both microscopes are equipped with a K2 Summit electron detector (Gatan, lnc.), and the data were collected using a beam image-shift data collection method40. The datasets were collected under a defocus range between –1.2 and –1.7 μm with a pixel size of 1.32 Å for the Talos Arctica microscope and 1.36 Å for the Titan Krios G2 microscope using SerialEM41. Each movie stack was dose fractionated into 32 movie frames with a total dose of 50 e– per Å2.
Cryo-EM data processing
For all cryo-EM datasets, beam-induced image motion of each movie stack was corrected using MotionCor242, and the parameters of the contrast transfer function (CTF) were estimated using CTFFIND443 on the aligned micrographs without dose weighting. The dose-weighted micrographs were used for further data processing using RELION 344 and a script developed for the block-based reconstruction45. For the EEEV PE6 VLP-LA1-8-Fc dataset, a small subset of about 500 particles was manually picked and processed using reference-free 2D classification. Three 2D class-averaged images were selected as references for particle auto-picking of the entire dataset, generating a total of 59,643 particles from 9215 micrographs. The particles were selected through several rounds of 2D classifications, which resulted in a set of 20,107 particles. The particles were further subjected to 3D classifications using the density map of EEEV14 low-passed to 15 Å as the initial reference. Finally, 14,691 particles from two classes with clear features were kept for the subsequent 3D auto-refinements with icosahedral symmetry imposed, yielding a density map of the EEEV PE6 VLP/VLDLR LA1-8-Fc complex with an overall resolution of 5.3 Å. To further improve the resolution of the density map, the block-based reconstruction method45 was applied to the refined particles. Blocks around the asymmetric unit were generated and re-extracted from the micrographs. After 3D classifications without image alignment, 298,035 particles were selected for further 3D auto-refinements, resulting in a 3.5 Å density map of the block that contains one asymmetric unit of the particle. The resolutions were measured according to the gold-standard Fourier shell correlation (FSC) criterion of a 0.143. The local-resolution maps were calculated by using ResMap46.
Similar data processing steps were performed for other datasets (Supplementary Tables 1, 2). Briefly, for the dataset of the EEEV PE6 VLP-VLDLR LA1-2-Fc complex, a total of 15,654 particles were selected for the final 3D auto-refinement with icosahedral symmetry imposed, resulting in a density map with an overall resolution of 5.2 Å. For the block-based reconstruction45, 350,983 block particles were selected and the reconstruction with the blocks generated a density map at a resolution of 3.4 Å after 3D auto-refinements. For the dataset of the EEEV PE6 VLP-VLDLR LA3-5-Fc complex, 14,840 particles were selected for the final 3D auto-refinement with icosahedral symmetry imposed, which produced a density map at a resolution of 4.8 Å. Further block-based reconstructions yielded a density map at a resolution of 3.4 Å. For other complexes, the datasets were processed by following a similar procedure as described above (Supplementary Table 1).
Model building and refinement
Model building was initiated by using the structures of EEEV E1, E2, and capsid proteins (pdb accession number: 6XO4)14, and the AlphaFold structures32 as references47. The models were manually adjusted in COOT48 and refined iteratively by real-space refinement in Phenix49. MolProbity50 was used to validate the geometries of the structures. The cryo-EM data collection, refinement, and validation statistics are summarized in the Supplementary Table 1. Some of the figures were prepared by using the UCSF chimera47 and chimeraX51.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and EM maps have been deposited into the Protein Data Bank (http://www.pdb.org) and the EM Data Bank, respectively. The deposited data include an asymmetric unit of the EEEV PE6 /LA1-2 complex (PDB-8XI4, EMD-38370), the intact EEEV PE6/LA1-2 complex (PDB-8YS2, EMD-38376), an asymmetric unit of the EEEV PE6/LA3-5 complex (PDB-8XI5, EMD-38371), the intact EEEV PE6/LA3-5 complex (PDB-8YS4, EMD-38377), EEEV PE6/LA1-8 (EMD-38378 [https://www.ebi.ac.uk/emdb/EMD-38378]), EEEV PE6/ApoERiso2 LA1-3 (EMD-38379 [https://www.ebi.ac.uk/emdb/EMD-38379]), EEEV PE6/LA1-3 (EMD-38380 [https://www.ebi.ac.uk/emdb/EMD-38380]), EEEV PE6/LA2-3 (EMD-38381 [https://www.ebi.ac.uk/emdb/EMD-38381]), EEEV PE6/LA1-6-W132A (EMD-38382 [https://www.ebi.ac.uk/emdb/EMD-38382]), EEEV PE6-K156A/LA1-8 (EMD-38383 [https://www.ebi.ac.uk/emdb/EMD-38383]), EEEV PE6-K206E/LA1-8 (EMD-38384 [https://www.ebi.ac.uk/emdb/EMD-38384]). The source data underlying Figs. 2, 4, 5, Supplementary Figs. 3, 6, 11 and 13 are provided as a Source Data file. Specific data P values are also included within Source Data file. Source data are provided with this paper.
References
Lindsey, N. P., Martin, S. W., Staples, J. E. & Fischer, M. Notes from the Field: multistate outbreak of eastern equine encephalitis virus - United States, 2019. MMWR Morb. Mortal. Wkly Rep. 69, 50–51 (2020).
de Novaes Oliveira, R. et al. Eastern equine encephalitis cases among horses in Brazil between 2005 and 2009. Arch. Virol. 159, 2615–2620 (2014).
Lindsey, N. P., Staples, J. E. & Fischer, M. Eastern equine encephalitis virus in the United States, 2003–2016. Am. J. Tropical Med. Hyg. 98, 1472–1477 (2018).
Armstrong, P. M. & Andreadis, T. G. Eastern equine encephalitis virus in mosquitoes and their role as bridge vectors. Emerg. Infect. Dis. 16, 1869–1874 (2010).
Sherwood, J. A., Stehman, S. V., Howard, J. J. & Oliver, J. Cases of Eastern equine encephalitis in humans associated with Aedes canadensis, Coquillettidia perturbans and Culiseta melanura mosquitoes with the virus in New York State from 1971 to 2012 by analysis of aggregated published data. Epidemiol. Infect. 148, e72 (2020).
Sah, R. et al. The emerging scenario for the Eastern equine encephalitis virus and mitigation strategies to counteract this deadly mosquito-borne zoonotic virus, the cause of the most severe arboviral encephalitis in humans—an update. Front. Tropical Dis. 3, 1077962 (2023).
Aréchiga-Ceballos, N. & Aguilar-Setién, A. Alphaviral equine encephalomyelitis (Eastern, Western and Venezuelan). Rev. Sci. Tech. 34, 491–501 (2015).
Arrigo, N. C., Adams, A. P. & Weaver, S. C. Evolutionary patterns of eastern equine encephalitis virus in North versus South America suggest ecological differences and taxonomic revision. J. Virol. 84, 1014–1025 (2010).
Ciota, A. T. Eastern equine encephalitis virus taxonomy, genomics, and evolution. J. Med Entomol. 59, 14–19 (2022).
Kim, A. S. & Diamond, M. S. A molecular understanding of alphavirus entry and antibody protection. Nat. Rev. Microbiol. 21, 396–407 (2023).
Holmes, A. C., Basore, K., Fremont, D. H. & Diamond, M. S. A molecular understanding of alphavirus entry. PLoS pathog. 16, e1008876 (2020).
Firth, A. E., Chung, B. Y. W., Fleeton, M. N. & Atkins, J. F. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol. J. 5, 108 (2008).
Sutton, M. S. et al. Vaccine elicitation and structural basis for antibody protection against alphaviruses. Cell 186, 2672–2689.e25 (2023).
Williamson, L. E. et al. Human antibodies protect against aerosolized eastern equine encephalitis virus infection. Cell 183, 1884–1900.e23 (2020).
Chen, C.-L. et al. Cryo-EM structure of eastern equine encephalitis virus in complex with heparan sulfate analogues. Proc. Natl Acad. Sci. USA 117, 8890–8899 (2020).
Hasan, S. S. et al. Cryo-EM structures of eastern equine encephalitis virus reveal mechanisms of virus disassembly and antibody neutralization. Cell Rep. 25, 3136–3147.e5 (2018).
Clark, L. E. et al. VLDLR and ApoER2 are receptors for multiple alphaviruses. Nature 602, 475–480 (2022).
Ma, H. et al. LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 588, 308–314 (2020).
Zimmerman, O. et al. Vertebrate-class-specific binding modes of the alphavirus receptor MXRA8. Cell 186, 4818–4833.e25 (2023).
Zhang, R. et al. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574 (2018).
Cao, Y., Wang, H., Jin, P., Ma, F. & Zhou, X. Identification and characterization of the very-low-density lipoprotein receptor gene from branchiostoma belcheri: insights into the origin and evolution of the low-density lipoprotein receptor gene family. Anim. (Basel) 13, 2193 (2023).
Nimpf, J. & Schneider, W. J. From cholesterol transport to signal transduction: low density lipoprotein receptor, very low density lipoprotein receptor, and apolipoprotein E receptor-2. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1529, 287–298 (2000).
Fass, D., Blacklow, S., Kim, P. S. & Berger, J. M. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388, 691–693 (1997).
Verdaguer, N., Fita, I., Reithmayer, M., Moser, R. & Blaas, D. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11, 429–434 (2004).
Ma, B., Huang, C., Ma, J., Xiang, Y. & Zhang, X. Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3. Nature 598, 677–681 (2021).
Basore, K. et al. Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor. Nature 598, 672–676 (2021).
Cao, D., Ma, B., Cao, Z., Zhang, X. & Xiang, Y. Structure of Semliki Forest virus in complex with its receptor VLDLR. Cell 186, 2208–2218.e15 (2023).
Song, H. et al. Molecular basis of arthritogenic alphavirus receptor MXRA8 binding to chikungunya virus envelope protein. Cell 177, 1714–1724(2019).
Basore, K. et al. Cryo-EM structure of chikungunya virus in complex with the Mxra8 receptor. Cell 177, 1725–1737. e16 (2019).
Adams, L. J. et al. Structural and functional basis of VLDLR usage by Eastern equine encephalitis virus. Cell 187, 360–374.e19 (2024).
Platteborze, P. L., Kondig, J. P., Schoepp, R. J. & Wasieloski, L. P. Comparative sequence analysis of the eastern equine encephalitis virus pathogenic strains FL91-4679 and GA97 to other North American strains. DNA Seq. 16, 308–320 (2005).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Octet® BLI Label-Free Detection Systems for Biomolecular Interactions Analysis. (2023).
Tiebel, O. et al. Mouse very low-density lipoprotein receptor (VLDLR): gene structure, tissue-specific expression and dietary and developmental regulation. Atherosclerosis 145, 239–251 (1999).
Maire, L. F., 3rd, McKinney, R. W. & Cole, F. E., Jr. An inactivated eastern equine encephalomyelitis vaccine propagated in chick-embryo cell culture. I. Production and testing. Am. J. Trop. Med. Hyg. 19, 119–122 (1970).
Bartelloni, P. J., McKinney, R. W., Duffy, T. P. & Cole, F. E. Jr. An inactivated eastern equine encephalomyelitis vaccine propagated in chick-embryo cell culture. II. Clin. serologic responses man. Am. J. Trop. Med Hyg. 19, 123–126 (1970).
Sakai, K. et al. A neuronal VLDLR variant lacking the third complement-type repeat exhibits high capacity binding of apoE containing lipoproteins. Brain Res 1276, 11–21 (2009).
Ko, S. Y. et al. A virus-like particle vaccine prevents equine encephalitis virus infection in nonhuman primates. Sci. Transl. Med 11, eaav3113 (2019).
Huang, X. et al. Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation. Prog. Biophysics Mol. Biol. 156, 3–13 (2020).
Wu, C., Huang, X., Cheng, J., Zhu, D. & Zhang, X. High-quality, high-throughput cryo-electron microscopy data collection via beam tilt and astigmatism-free beam-image shift. J. Struct. Biol. 208, 107396 (2019).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zhu, D. et al. Pushing the resolution limit by correcting the Ewald sphere effect in single-particle Cryo-EM reconstructions. Nat. Commun. 9, 1552 (2018).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Williams, C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
Cryo-EM data collection was carried out at the Center for Biological Imaging, Core Facilities for Protein Science at the Institute of Biophysics (IBP), Chinese Academy of Sciences (CAS). We thank L. Zhang, X. Huang, B. Zhu, L. Chen, and other staff members at IBP, CAS. We thank Xing Jia and Qing Bian from Center for Biological Imaging (CBI), Institute of Biophysics, Chinese Academy of Sciences, for technical support with LSM980 Confocal-Plus imaging and image analysis. YX, XZZ and DFC are funded by the National Natural Science Foundation of China (grants: 31930069, 31925023, 21827810, 31861143027, 32325027, 32371278, 32241027), the Ministry of Science and Technology of China (grants: 2023YFC2306300, 2021YFA1300204, 2021YFA1301501, 2017YFA0504700), the Tsinghua University Vanke Special Fund for Public Health and Health Discipline Development (NO. 2022Z82WKJ013), the Beijing Frontier Research Center for Biological Structure, the Tsinghua-Peking Center for Life Sciences, the Strategic Priority Research Program of the CAS (XDB37040101), the Key Research Program of Frontier Sciences at the CAS (ZDBS-LY-SM003), Basic Research Program Based on Major Scientific Infrastructures, CAS (JZHKYPT-2021-05), and the Key Laboratory of Biomacromolecules, Chinese Academy of Sciences (ZGD-2023-05). DFC was sponsored by the Beijing Nova Program (20230484239). BTM is supported by the China Postdoctoral Science Foundation (2023M731981 and 2023T160354).
Author information
Authors and Affiliations
Contributions
B.T.M. and X.Y.X. planned and performed the biochemical experiments, analyzed data, prepared the figures, and wrote the manuscript together with Y.X. and X.Z.Z. D.F.C. performed cryo-EM structure determination and analysis, prepared the figures, and wrote the manuscript together with Y.X. and X.Z.Z. Z.Y.C. preformed cryo-EM data collection, cell localization and attachment assay. Y.X. and X.Z.Z. planned and supervised the experiments, analyzed the data, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cao, D., Ma, B., Cao, Z. et al. The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes. Nat Commun 15, 6866 (2024). https://doi.org/10.1038/s41467-024-51293-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51293-x
- Springer Nature Limited