Abstract
Oligoclonal mixtures of broadly-neutralizing antibodies can neutralize complex compositions of similar and dissimilar antigens, making them versatile tools for the treatment of e.g., infectious diseases and animal envenomations. However, these biotherapeutics are complicated to develop due to their complex nature. In this work, we describe the application of various strategies for the discovery of cross-neutralizing nanobodies against key toxins in coral snake venoms using phage display technology. We prepare two oligoclonal mixtures of nanobodies and demonstrate their ability to neutralize the lethality induced by two North American coral snake venoms in mice, while individual nanobodies fail to do so. We thus show that an oligoclonal mixture of nanobodies can neutralize the lethality of venoms where the clinical syndrome is caused by more than one toxin family in a murine challenge model. The approaches described may find utility for the development of advanced biotherapeutics against snakebite envenomation and other pathologies where multi-epitope targeting is beneficial.
Similar content being viewed by others
Introduction
By definition, broadly-neutralizing antibodies can neutralize several similar antigens that are not identical but cause the same physiological effects and may therefore be useful for the treatment of many infectious diseases1,2,3, but certainly also snakebite envenomations, where multiple similar (and dissimilar) toxins in a venom need to be neutralized. In the Americas, over 2400 people fall victim to coral snake envenomations annually4. This group of snakes is highly diverse, and different species possess venoms that significantly differ in their overall composition5. However, the key targets to neutralize for therapeutic antibodies are the toxins relevant for the envenomation of mammals, including humans. Across coral snake venoms, these are relatively few and belong to only two protein subfamilies, namely neurotoxic phospholipases A2 (PLA2s) and α-neurotoxins (αNTxs) from the three-finger toxin family (3FTxs)5,6,7. When these toxins are injected into mammalian prey and victims, they exert neurotoxic effects that manifest clinically as flaccid paralysis of skeletal muscles, which, if left untreated, can be fatal as this condition may progress to respiratory failure8,9. Currently, the only available specific treatments for envenomed patients are antivenoms that consist of polyclonal antibodies isolated from the plasma of hyperimmunized animals10. While these antivenoms have saved countless lives, they unfortunately suffer from several drawbacks, including a limited capacity to cross-neutralize venoms from different coral snake species9,11, batch-to-batch variation, and a low content of therapeutically active antibodies12,13. In comparison with many other antivenoms, such as those for viper envenomations, the issue with the low amount of neutralizing antibodies is particularly relevant for coral snake antivenoms, as the low abundance and limited immunogenicity of some of the medically most relevant toxins in coral snake venoms (i.e., αNTxs)14,15 make it difficult to raise neutralizing antibodies via the animal immunization process used for traditional antivenom manufacturing15,16. Therefore, very high doses of antivenom are typically needed to treat severe envenomations, which further increases the risk of adverse reactions due to the heterologous nature of the antivenom antibodies.
To address the abovementioned issues, several researchers aim towards developing new types of antivenom products, e.g., recombinant antivenoms. One approach that has proven promising is the generation of recombinant antibodies against key venom toxins using phage display technology17,18,19. This methodology enables the discovery of specific antibodies against the most medically relevant toxins, regardless of their abundance within the venoms or immunogenicity20. Phage display technology can furthermore facilitate the discovery of cross-neutralizing antibodies (antibodies that can neutralize more than one toxin isoform) through cross-panning strategies17,19,21 or selections against recombinantly produced consensus antigens (i.e., antigens designed to represent the ‘average’ of several different proteins)22. Finally, it has been shown that monoclonal antibodies can be combined as carefully generated oligoclonal mixtures, allowing for the neutralization of multiple toxins by a single cocktail18. These discoveries have further led to the speculation that recombinant antivenoms with very broad neutralization capacity (i.e., polyvalent recombinant antivenoms) can be developed by preparing oligoclonal mixtures of individual cross-neutralizing monoclonal antibodies23 and that this may be a promising approach to develop a new type of affordable envenomation therapies24,25.
Replacing polyclonal antibodies purified from the plasma of immunized animals with recombinant oligoclonal antibody mixtures may have the potential to significantly reduce batch-to-batch variation, to neutralize all medically relevant toxins in the targeted venoms, and to ensure a high therapeutic antibody content in the recombinant products. To date, work on recombinant monoclonal antibodies has primarily focused on using human monoclonal immunoglobulin G (IgG) antibodies, which are highly specific and have long half-lives in circulation, but have limited stability ex vivo and are relatively expensive to manufacture compared to other types of recombinant binding proteins25. As an alternative, other therapeutically promising antibody scaffolds, such as camelid single-domain antibodies (VHHs, also known as nanobodies) have come into focus26. VHHs are derived from heavy-chain-only antibodies present in Camelidae and are characterized by possessing similarly high affinities and specificities as IgG antibodies. They are more stable at high temperatures and extreme pH27 than IgGs, and they can generally be expressed in a large scale at a lower cost25. Their small size (12–15 kDa) increases their ability to penetrate deep tissues but has the drawback of resulting in a short serum half-life. However, if needed, the circulation half-life can be optimized through protein engineering techniques, such as fusion with a human Fc domain26,28 or assembly into larger protein architectures19,29 (although seldom without affecting other parameters such as tissue penetration).
To investigate the utility of VHHs against snakebite envenomation, in this study, we aimed to discover cross-neutralizing VHHs against the key toxins in coral snake venoms to enable the preparation of an oligoclonal VHH mixture that could be used to treat coral snake envenomation. To this end, we used an immune VHH phage display library from one alpaca and one llama immunized with multiple elapid snake venoms for discovery of VHHs targeting the medically most important toxins in coral snake venoms. To further facilitate the discovery of cross-neutralizing VHHs, we utilized a recombinant consensus antigen representing αNTxs and a native representative neurotoxic PLA2 as antigens. Using both a rodent model involving pre-incubation of venom and VHHs, as well as a rodent model mimicking a real-life snakebite envenomation, where venom is first injected subcutaneously (s.c.), followed by administration of VHHs intravenously (i.v.) (a.k.a. a rescue model), we demonstrated that the discovered VHHs can neutralize the lethality of coral snake neurotoxins. Furthermore, we showed that an oligoclonal mixture of only two cross-neutralizing VHHs, even in a monovalent format, can neutralize the lethality of the whole venoms of Micrurus fulvius (Eastern coral snake, US) and Micrurus diastema (variable coral snake, Mexico) performing comparably to the existing plasma derived antivenom (Coralmyn), while individual VHHs fail to do so on their own. Finally, we explore the utility of two alternative antibody constructs (a homodimeric VHH-VHH construct and a VHH-Fc construct) and conclude that these do not perform better than the simpler monovalent VHHs.
Results
Camelid immunization and VHH phage display library generation
To facilitate the discovery of cross-neutralizing VHHs, one alpaca and one llama were immunized with increasing doses of a mixture of 18 elapid venoms over 16-weeks (Supplementary Table 1), followed by construction of VHH displaying phage libraries. Blood samples were collected from the animals before immunization (day 0) and preceding venom inoculation on each injection day. Subsequently, an Enzyme-Linked Immunosorbent Assay (ELISA) was employed to analyze the antibody responses over time against each of the individual venoms used for immunization. Antibody binding signals increased over time for all venoms, and both camelids showed comparable immune responses against the same venoms. For example, lower binding signals were observed against Dendroaspis venoms compared to Naja venoms (Fig. 1). After 8 and 16 weeks of immunization, one VHH-displaying phage library was generated from each animal, all larger than 3·108 individual clones with insert rates > 85%. As serum samples from the two animals showed a comparable immune response, the phage libraries were mixed, resulting in one library from 8 weeks and one from 16 weeks of immunization.
Antibody binding signals observed for serum samples collected at different time points from two camelids immunized with a mixture of 18 elapid venoms. A Response of llama 0406 to the 18 venoms included in the immunization mixture. B Response of alpaca 0541 to the 18 venoms included in the immunization mixture. Values correspond to the means of two replicates (n = 2). The signal at day 0 represents the response of the preimmune sera. Source data are provided as a Source Data file.
Phage display selection campaigns and screening of VHH binders
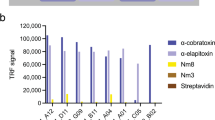
To enable the preparation of an oligoclonal cocktail for the treatment of coral snake envenomation, we focused our discovery efforts of VHHs against the medically most relevant toxins in coral snake venoms namely neurotoxic PLA2s, from the PLA2 family, and αNTxs from the 3FTx family. The decision to use this immune phage display library to discover VHHs against coral snake venom toxins was based on the high sequence similarity observed between toxins in coral snake venoms and in venoms from other elapid snake species such as Naja and Dendroaspis. An alignment of short chain αNTxs from these genera can be found in Supplementary Fig. 1. The 16-week library was panned against purified toxins selected to facilitate the discovery of cross-neutralizing VHHs. These included two wildtype PLA2s purified from the venom of M. fulvius (denominated PLA2N and PLA2O6), two recombinantly expressed native αNTxs from the venoms of Micruroides euryxanthus and M. diastema (named rEury30 and rDH31, respectively), and a recombinantly expressed short-chain consensus αNTx (named scNTx32). After two or three consecutive rounds of selection, five enriched libraries (TPL0637, TPL0638, TPL0622, TPL0623, and TPL0629) were chosen for further studies based on the enrichment of the phage pools (i.e., the number of colony-forming units in the performed selection outputs compared to a negative selection run in parallel without any antigen) (Supplementary Figs. 3A and 4A). The selected libraries were subcloned into a modified pHEN6 expression vector for soluble VHHs and transformed into BL21 (DE3) cells33. Monoclonal VHHs from individually picked clones were expressed and binding to their cognate target was assessed using an expression-normalized capture Dissociation-Enhanced Lanthanide Immunoassay (DELFIA) (Supplementary Figs. 2B, C and 3B–D). In agreement with the enrichment of the phage pools, a higher fraction of binding clones was observed from the selection campaign performed using the consensus scNTx (Supplementary Fig. 3D) compared to the campaigns using native toxins as antigens (Supplementary Fig. 3B, C). This could indicate that the epitope(s) of the consensus toxin have more in common with the epitope(s) of the toxins used for immunization compared to the native coral snake toxins used in the experiment (rEury and rDH). Therefore, in this experiment, the use of this consensus toxin was advantageous over the use of the native toxins. Based on the screening results, 24 and 49 clones with high binding signals to PLA2 and αNTxs, respectively, were selected for sequencing. Out of the 79 sequenced clones, 19 anti-PLA2 and 17 anti-αNTx VHHs showed unique sequences, which could be further clustered in 4 and 7 families, respectively, based on sequence similarity of the complementarity-determining regions (CDRs).
The three unique VHH clones targeting either PLA2s or αNTxs (Supplementary Fig. 5) that showed the highest binding signals in the expression-normalized capture DELFIA were further analyzed in dose-response experiments to assess cross-reactivity to PLA2s and αNTxs from other snake genera. The three anti-PLA2 VHHs all showed comparable binding to PLA2N- and PLA2-containing fractions from the venoms of Naja melanoleuca (Nm15), Naja nigricollis (Nn19), and Hemachatus haemachatus (Hh3) with TPL0638_01_C09 showing the highest binding signal. No binding to a structurally different PLA2 from Echis pyramidum was observed (Supplementary Fig. 5). Similarly, the three αNTx-targeting VHHs also bound to αNTxs from the venoms of M. diastema (DH), Naja haje (Nh1), Dendroaspis viridis (Dv1), and H. haemachatus (Hh1) with TPL0629_01_D11 showing the highest binding signal and the lowest EC50-value (Supplementary Fig. 6). No binding to α-cobratoxin, a long chain αNTx from the 3FTx family purified from the venom of Naja kaouthia, was observed. Taken together, the high affinity of the discovered VHHs to PLA2s and αNTxs from different snake genera demonstrates that they possess cross-reactive binding across their respective toxin (sub)families.
Analysis of VHH binding kinetics with biolayer interferometry
To characterize the binding kinetics of the selected VHH clones against each toxin group, biolayer interferometry (BLI) experiments were performed using PLA2N and αNTx DH as antigens. The data were fitted to a 1:1 binding model, which assumes a single VHH molecule binds to a single toxin molecule. All the tested VHHs showed high affinity, with KD values in the pM range for the three anti-PLA2 VHHs and in the nM range for the three anti-αNTx VHHs. Four of the six monovalent VHHs showed very slow dissociation rates (koff < 5.5·10−4 s−1), which indicates that they remain bound to their respective toxin for a long time period (Fig. 2, Table 1).
Biotinylated toxins captured on streptavidin biosensors were dipped in decreasing concentrations of each of the VHHs, followed by dissociation in kinetics buffer. Binding data were fitted using a 1:1 model. The colors represent the different VHH concentrations: black is 200 nM, pink is 67 nM, green is 22 nM, dark purple is 7.4 nM, light purple is 2.5 nM, and cyan is 0.8 nM. A–C Anti-PLA2 VHHs binding PLA2N. D–F Anti-αNTx VHHs binding αNTx DH. Source data are provided as a Source Data file.
Neutralization of PLA2 enzymatic activity
To assess the potential cross-neutralizing capacity of the PLA2-binding VHHs, an in vitro enzymatic PLA2 activity neutralization assay was performed using a commercial kit and three of the PLA2s and PLA2-containing fractions that the VHHs showed binding to (PLA2N, Nn19, and Hh3). The VHHs were evaluated at a 1:20 toxin to VHH molar ratio (0.07 µM and 1.4 µM). All three anti-PLA2 VHHs reduced the enzymatic activity of PLA2N as well as the PLA2 from the venom of N. nigricollis (Nn19) but showed no inhibition of the activity of Hh3 (H. haemachatus) (Fig. 3A). The VHHs alone did not show any PLA2 activity. This result demonstrates the neutralization capacity of the VHHs for PLA2s beyond those in the venoms of Micrurus. Further work is necessary to assess the utility of these VHHs to neutralize the toxic effects of H. haemachatus PLA2s, as well as PLA2s from other snake genera.
A Inhibition of PLA2 enzymatic activity by VHHs. The maximal enzyme activity observed with toxin alone was set to 100%. The normalized enzymatic activity of PLA2s from various elapid genera (PLA2N, Nn19, and Hh3) preincubated for 30 min at RT with anti-PLA2 VHHs at a 1:20 toxin to VHH molar ratio. Bars represent the mean of two replicates. B Neutralization of αNTx-mediated blocking of muscle-type nAChR current. Dose-response curves from patch clamp experiments with increasing concentrations of VHHs to prevent the blocking of nAChR by 15 nM αNTx DH or 5 nM scNTx. Error bars represent standard deviation of independent cells performed in a 384-well plate. n = 16. Source data are provided as a Source Data file.
Neutralization of αNTx-mediated blocking of the nicotinic acetylcholine receptor (nAChR)
The ability of the discovered anti-αNTx VHHs to neutralize αNTx-mediated blocking of the muscle type nAChR function was evaluated using whole-cell patch clamp recordings of rhabdomyosarcoma cells endogenously expressing the nAChR, which were exposed to purified αNTx DH or the consensus toxin scNTx at a concentration resulting in approximately 80% inhibition (IC80) (Fig. 3B). When tested against the αNTx DH, two of the VHHs could completely neutralize the αNTx activity at VHH concentrations higher than 45 nM, corresponding to molar ratios above 1:3 between the toxin and VHH. Approximately half of the toxins’ activity was neutralized at a 1:1 toxin to VHH molar ratio (corresponding to a VHH concentration of 15 nM). The three VHHs performed better against the consensus toxin scNTx where full neutralization was observed at a 1:1 toxin to VHH molar ratio (5 nM of VHH) for two of the VHHs.
In vivo neutralization of purified toxins
Based on the in vitro binding and neutralization data, the three most promising VHHs against PLA2s and αNTxs were assessed for neutralization of lethality with purified toxins via intravenous (i.v.) injection of the preincubated toxin and VHH (preincubation experiments) and i.v. injection of VHH after envenomation using subcutaneous (s.c.) injection (rescue experiments). The VHHs were evaluated for their ability to neutralize PLA2N and wild-type αNTx DH induced lethality, respectively. As negative controls, two groups of mice were injected with 3 median lethal doses (LD50s) of PLA2N or αNTx DH preincubated with an isotype VHH. As expected, no neutralization or delay of time of death was observed in these groups. Also, a group of mice was injected i.v. with the highest dose evaluated of each VHH. None of these mice showed any signs of adverse reactions (in vivo data are shown in Supplementary Tables 2 and 3).
Neutralization of PLA2-induced lethality was evaluated using 3 LD50s of PLA2N, purified from the venom of M. fulvius. The LD50 of this toxin was determined to be 10.3 µg/mouse when given i.v. and 34.6 µg/mouse with s.c. administration. The three anti-PLA2 VHHs completely neutralized PLA2N-induced lethality when preincubated with PLA2N and injected i.v. at a 1:1 toxin to VHH molar ratio (Fig. 4A). Conversely, when the VHHs were injected i.v. immediately after s.c. injections of PLA2N (1:2.5 toxin to VHH molar ratio), only TPL0637_01_A07 neutralized PLA2N-induced lethality in all three mice, while the other two rescued 1 of the 3 mice (Fig. 4B).
A, C 3 LD50s of PLA2N (A) or αNTx DH (C) were preincubated with either PBS or one of the VHHs and injected in mice using the i.v. route. n = 3. B, D The mice were injected with 3 LD50s of PLA2N (B) or αNTx DH (D) using the s.c. route followed by immediate i.v. injection with PBS or one of the VHHs. Toxin to VHH molar ratio is presented in parentheses. n = 3. * Indicates a significant difference to PBS control (P < 0.05) in a Mantel-Cox log-rank test. Source data are provided as a Source Data file.
Even though αNTxs are in relatively low abundance in Mexican coral snake venoms, it has been shown, using polyclonal sera from immunized horses, that the presence of neutralizing antibodies against these toxins is necessary for the neutralization of several of these venoms, including M. diastema14. The LD50 of wild-type αNTx DH from the venom of M. diastema was determined to be 2.0 µg/mouse when given i.v. and 4.8 µg/mouse with s.c. administration and neutralization of lethality by VHHs was evaluated using 3 LD50s of the toxin. TPL0629_01_D11 and TPL0629_01_A07 prevented lethality in all the mice when preincubated with toxin at a 1:2.5 molar ratio between toxin and VHH followed by i.v. injection of the mixture (Fig. 4C). In the same type of experiment, TPL0629_01_G06 did not prevent death of mice using the same molar ratio between toxin and VHH. Although survival of the mice was identical for the first two VHHs, some signs of paralysis of the mice were observed when using TPL0629_01_A07 for neutralization and therefore the following experiments were only performed with TPL0629_01_D11. When 3 LD50s of the toxin were injected s.c. followed by immediate i.v. injection of the VHH, all mice were rescued using a 1:10 toxin to VHH molar ratio (Fig. 4D). To evaluate if a bivalent version of TPL0629_01_D11 would allow for a lower molar excess of the VHH to be used for neutralization, two new constructs, a bivalent VHH construct, where two VHHs are connected by a GS-linker, and a VHH-Fc, in which the VHH is fused to a human Fc domain, were produced and evaluated in vivo. To confirm that both binding sites of the bivalent constructs are available for antigen binding, the constructs were analyzed in BLI with immobilized toxin as described in Materials and Methods (Section 4.10). Both constructs showed an increased avidity for αNTx DH (0.53 nM for the bivalent VHH and 3.69 nM for the VHH-Fc) compared to the monovalent VHH (6.94 nM) (Supplementary Fig. 7). For neutralization, a 1:1.25 molar ratio between toxin and VHH-Fc or the bivalent VHH construct (corresponding to the same ratio between toxin and binding sites for all evaluated constructs) was used. The VHH and the VHH-Fc showed similar results, delaying the time of death for about 8–10 h, while the bivalent VHH construct only prolonged the survival to about 3 h (Fig. 5). As no benefit of using the bivalent constructs was observed, further work focused only on the monovalent VHH construct.
The mice were injected with 3 LD50s of αNTx DH using the s.c. route followed by immediate i.v. injection with PBS, or the VHH TPL0629_01_D11 as a monovalent VHH construct, and as bivalent constructs (bivalent VHH, or VHH-Fc). Toxin to antibody construct molar ratios are presented in parentheses. Note that a molar ratio of 1:1.25 of the bivalent constructs is equivalent to a 1:2.5 molar ratio between toxin and binding sites. n = 3. * Indicates a significant difference to PBS control (P < 0.05) in a Mantel-Cox log-rank test. Source data are provided as a Source Data file.
In vivo neutralization of whole coral snake venoms
As previously mentioned, PLA2s and αNTxs are determined to be the key targets in the venom of coral snakes. Based on the work of Vergara et al.6, approximately 60% of the venom of M. fulvius is composed of PLA2s and around 32% of 3FTxs (Supplementary Fig. 8). RP-HPLC analysis of the venom of M. diastema has shown a similar venom composition, with approximately 62% of the total protein content being PLA2s and 22% 3FTxs (Supplementary Fig. 8). Here, it is relevant to note that toxins in these venoms are not limited to PLA2N and αNTx DH, but the venoms also contain other similar toxins that need to be neutralized to prevent complete venom-induced lethality. To prepare oligoclonal mixtures containing a minimal, but sufficient, number of VHHs, the most potent of the discovered VHHs, namely TPL0629_01_D11 (neutralizing αNTxs) and TPL0637_01_A07 (neutralizing PLA2s) were included in two different mixtures. The molar ratios of the VHHs in these mixtures were based on the molar ratio between PLA2s and 3FTxs in the venoms (Supplementary Table 4). Thereafter, the oligoclonal mixtures were evaluated for their ability to neutralize the whole venoms from M. fulvius and M. diastema.
The LD50s of the venoms were determined to be 6.0 µg/mouse for M. fulvius and 5.7 µg/mouse for M. diastema, using the i.v. route for administration. When 3 LD50s of each venom were preincubated with each oligoclonal mixture using a 1:10 toxin to VHH molar ratio prior to i.v. administration, the respective mixture was able to prevent lethality in all the mice injected with M. fulvius venom and two of the three mice injected with M. diastema venom. For comparison, a plasma-derived antivenom used for the treatment of coral snake envenomation in Mexico, Coralmyn, was included as a control at a similar dose as the oligoclonal mixtures (a 1:10 ratio between toxin and antibody binding sites). Coralmyn prevented lethality in all mice injected with M. fulvius venom, whereas the three mice injected with M. diastema venom died within 3 h. As a reference, the final doses of the oligoclonal mixtures used were 14.0 mg/kg for M. fulvius and 12.9 mg/kg for M. diastema compared to 45.5 mg/kg and 40.7 mg/kg for Coralmyn. Preincubation of both M. fulvius or M. diastema venom with either TPL0637_01_A07 (targeting PLA2s) or TPL0629_01_D11 (targeting αNTxs) alone did not prevent lethality or prolong survival for any of the envenomed mice (Fig. 6). Overall, the data show that both of the oligoclonal mixtures possess a comparable neutralization capacity than the traditional antivenom, Coralmyn, on the two tested venoms, indicating a possibly broader species coverage (in vivo raw data is shown in Supplementary Table 5). Further optimization and neutralization assays on other coral snake venoms could, however, likely be used to identify an even more optimal VHH mixture for a final recombinant antivenom product.
3 LD50s of venom from (A). M. fulvius or (B). M. diastema, were preincubated with either PBS, the individual VHHs, the relevant VHH oligoclonal mixture prepared for the specific venom, or the polyclonal F(ab’)2-based antivenom, Coralmyn. Approximate toxin to VHH or F(ab’)2 molar ratios are shown in parentheses. Calculations are based on total protein content in the Coralmyn and oligoclonal mixtures. n = 3. * Indicates a significant difference to PBS control (P < 0.05). # Indicates a significant difference to Coralmyn (P < 0.05) in a Mantel-Cox log-rank test. Source data are provided as a Source Data file.
Discussion
The use of oligoclonal antibody mixtures to combat pathologies holds significant therapeutic promise, and such mixtures have already been tested against a number of indications, including several infectious diseases34,35,36. These advanced biologics derive their therapeutic potential from their ability to bind multiple targets or epitopes, which can be exploited to modulate intricate disease mechanisms or to neutralize several dissimilar pathogens. One of the indications for which oligoclonal antibody mixtures may be especially useful is snakebite envenomation. Here, complex mixtures of similar and dissimilar toxins need to be neutralized to prevent the onset of toxic effects37,38. To neutralize similar toxins from the same protein family, single cross-neutralizing monoclonal antibodies targeting variations of the same epitope can be applied19. However, developing therapies against snakebite envenomation that cover multiple snake species, let alone a whole venom from a single snake species (which often contains several different toxin families of medical importance), large benefits emerge by utilizing oligoclonal antibody mixtures. For these to be effective, multiple cross-neutralizing monoclonal antibodies should be combined into broadly-neutralizing oligoclonal antibody mixtures that can neutralize several entire (sub)families of toxins23. In this study, we developed a prototype mixture based on cross-neutralizing VHHs and demonstrated its ability to neutralize lethality induced by two North American coral snake venoms in a mouse model. Previous studies have shown that a single monoclonal antibody can neutralize the lethal effects of snake venom when lethality is caused by one predominant toxin39,40, that such antibodies can be further developed to become broadly-neutralizing19, and that oligoclonal antibodies can be used to neutralize multiple different toxins18. However, here, we go beyond the state of the art by demonstrating that the concepts from these previous studies can be merged and applied to a simpler antibody format, namely the VHH format, thereby providing a proof of concept for a different type of antivenom41. This work also serves as a proof of principle for creating cross-neutralizing, multi-epitope targeting oligoclonal antibody mixtures, which may find application beyond animal envenomations.
Specifically, in this study, we used existing data on the proteomic composition and toxicity of venoms from coral snakes5,6,7,42,43,44 to focus our discovery efforts on the medically relevant presynaptic-acting PLA2s and postsynaptic-acting αNTxs and establish a discovery approach for cross-neutralizing VHHs17,21. Using this approach, we discovered multiple VHHs and showed that three anti-αNTx VHHs bind a native αNTx from M. diastema with high affinity (Fig. 2, Table 1) as well as other αNTxs purified from the venoms of N. haje, D. viridis, and H. haemachatus (Supplementary Fig. 6), demonstrating their cross-reactive binding capabilities. Similarly, we showed that the three tested anti-PLA2 VHHs can neutralize the enzymatic activity of PLA2s from both Micrurus and Naja venoms in vitro, demonstrating their cross-neutralizing capacity (Fig. 3A). Not surprisingly, the discovered VHHs were unable to recognize a structurally different group of PLA2s purified from the venom of the viper E. pyramidum (Supplementary Fig. 5). In rescue experiments using a rodent model, which mimics a real-life snakebite scenario45, lethality induced by purified PLA2N or αNTx DH could be neutralized by three of the identified cross-reactive VHHs individually (Fig. 4). An evaluation of one of the VHHs in three different formats, as monovalent VHH, as a bivalent VHH construct, and as a VHH-Fc construct, for rescuing mice envenomed with αNTx revealed that, in this experimental setting, a larger bivalent antibody format did not exhibit any advantages in terms of prolonged survival compared to the simpler VHH format. While the exact reason for this remains unknown, we speculate that the complex toxicokinetic and pharmacokinetic interplay between the toxins and the VHH constructs plays a role. These findings, nevertheless, underscore the utility of VHHs for treating envenomation with venoms primarily composed of low molecular weight components, which have fast tissue absorption, such as PLA2s and 3FTxs46,47,48. Finally, we generated two oligoclonal mixtures using the best neutralizing VHH for each toxin family. We showed that a combination of only two VHHs could neutralize the lethality of complete coral snake venoms in vivo, but also that both VHHs were necessary to achieve this effect, as the two VHHs did not prevent lethality when administered individually (Fig. 5). Furthermore, we demonstrate that these two mixtures can cross-neutralize venoms from M. fulvius and M. diastema, unlike the antivenom that is currently used in the clinic (i.e., Coralmyn), which was only able to neutralize venom from M. fulvius (Fig. 6). In addition, the VHH mixtures can be used at lower doses in terms of mg/kg compared to Coralmyn. It is relevant to note that this comparison does not consider the percentage of venom-specific antibodies present in polyclonal antivenoms, such as Coralmyn, since this is unknown. Given the abundance of PLA2s and αNTxs similar to the ones used in this work in other North American coral snake venoms5,30,49,50, our results support the feasibility of generating and applying recombinant oligoclonal antivenoms, composed of only a few VHHs, for neutralizing venoms from coral snake species in North America. Eventually, we predict that this approach could be employed in the design of genus-wide recombinant antivenoms or even polyvalent recombinant antivenoms covering several snake genera.
While the work presented here has promise, it still has some limitations. For example, this study did not attempt the neutralization of complete coral snake venoms in a rescue setting due to the high VHH concentration required. To reduce the amount of VHHs needed for neutralization, future work should focus on the discovery of anti-αNTx VHHs with higher affinity to achieve neutralization at lower molar ratios similar to what has been seen previously with IgGs19,39. Our data showed that a higher affinity, specifically a slower dissociation rate between the VHH and αNTx, correlates with better neutralizing properties (Figs. 2 and 3, Table 1). In addition to the discovery of new VHHs with higher affinity, the current VHHs could potentially gain improved affinity through Bayesian optimization or random mutagenesis of the CDR regions51,52. Moreover, treatment of envenomation caused by more complex snake venoms, particularly those with high molecular weight enzymatic toxins, might require antibody formats with a longer circulation half-life, which can be achieved by leveraging different antibody scaffolds12. A final antivenom product could potentially combine various antibody formats and small molecule enzymatic inhibitors for optimal pharmacokinetic properties12,53. Finally, before any new type of antivenom product can be widely deployed, it will be paramount to establish how well the findings in this study and others translate to the clinical setting.
In the present study, we succeeded in discovering neutralizing VHHs against coral snake toxins from a VHH library made from a llama and an alpaca that were not immunized with coral snake venoms but only venoms from African cobras and mambas. The data presented here thus demonstrates that it is possible to discover para-specific VHHs with such a library, and we predict that it could also be used to discover VHHs against toxins from several other snake genera within the elapid snake family. If this hypothesis is correct, it indicates that the development of therapeutically beneficial recombinant antivenoms may be more facile than previously expected.
Beyond snakebite envenomation, the discovery pipeline presented here may find general application for the discovery and preparation of oligoclonal mixtures of cross-neutralizing VHHs against other protein targets involved in human diseases. We foresee that such antibody mixtures could find utility against bacterial, viral, and parasitic infections, where multiple virulence factors or targets must be neutralized. Conversely, oligoclonal mixtures of highly specific antibodies could also be applicable for disease targets where cross-reactivity of single antibodies could be detrimental due to high homology between the target antigen and endogenous non-target antigens in areas such as oncology and autoimmunity. In conclusion, our work demonstrates the feasibility of using oligoclonal VHH mixtures to neutralize complex snake venoms and shows the potential of discovering para-specific VHHs originally raised against toxins from other snake genera. Moreover, the discovery pipeline presented here can potentially be applied to address a range of other human diseases.
Methods
All animals and in vivo methodologies used in the present work were approved by the bioethics committee of the IBt-UNAM under project # 385 “Caracterización funcional y análisis de especificidad de venenos de coralillos Norteamericanos”. The Bioethics committee of the Instituto de Biotecnología, Universidad Nacional Autónoma de México (IBt-UNAM) is in compliance with the EU Directive 2010/63/EU for animal experiments54.
VHH phage display library generation
Immunization followed by generation of a VHH-displaying phage library targeting elapid snake venoms was commercially performed at the VIB nanobody core (Brussels, Belgium). For this, one alpaca and one llama were immunized with a mixture of venoms from 18 snake species (Dendroaspis angusticeps, D. jamesoni, D. polylepis, D. viridis, Naja anchietae, N. annulifera, N. ashei, N. haje, N. katiensis, N. melanoleuca, N. mossambica, N. nigricincta, N. nigricollis, N. nivea, N. nubiae, N. pallida, N. senegalensis, and Hemachatus haemachatus), including Gerbu adjuvant P as an adjuvant. The composition of the venoms included for immunization can be found in a study by Nguyen et al.55. Both camelids were injected s.c. bi-weekly at eight time points with increasing doses of the respective venom mixtures (See Supplementary Table 1 for the immunization scheme). Venoms were mixed, diluted in phosphate-buffered saline (PBS: 137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4.2H2O, 1.4 mM KH2PO4, pH 7.4) and mixed with Gerbu adjuvant P before injection.
To construct the VHH phage display libraries56, peripheral blood mononuclear cells (PBMCs) were isolated from the blood samples collected on days 46, 49, 102, and 105. The isolated PBMCs were used for total RNA extraction and libraries were prepared by pooling the total RNA samples after 46 and 49 days to generate a first library and after 102 and 105 days to generate a second library. These two pools of the total RNA samples were used as templates for first-strand synthesis of cDNA using oligo(dT) primers. Thereafter, VHH-encoding open reading frames were amplified by polymerase chain reaction (PCR), cloned into the phagemid vector pMECS, and transformed into electrocompetent E. coli TG1 cells.
Camelid antibody titer determination by enzyme-linked immunosorbent assay (ELISA)
White 96-well Immuno Plates (GR-655074, Thermo Fisher Scientific) were coated with 60 µL/well of the different whole venoms diluted in PBS (0.5 µg/mL) and incubated overnight at 4 °C. The next day, the plates were washed 4 times with PBST (PBS + 0.1% Tween 20), blocked with 200 µL/well of 0.5% bovine serum albumin (BSA) in PBST for 1 h at room temperature (RT), and washed 4 times with PBST. Then, 60 µL/well of plasma samples diluted to 0.4% (v/v) in 0.5% BSA-PBST were added and incubated for 1 h at RT, followed by 4 washes with PBST. Bound IgGs were detected with 60 µL/well of HRP-conjugated anti-alpaca IgG VHH domain (128-035-232, Jackson ImmunoResearch) diluted 1:10,000 in 0.5% BSA-PBST and incubated for 1 h at RT, followed by 4 washes with PBST. Finally, 60 μL/well of SuperSignal™ ELISA Pico Chemiluminescent Substrate (37070, Thermo Fisher Scientific) was added, and the plates were incubated for 5 min at RT before reading in a plate reader (VICTOR® Nivo™, PerkinElmer).
Venoms and purification of toxins
Venom from M. fulvius was obtained as a pool from 67 individual specimens, kindly donated by Jack Facente from “AGRITOXINS Venom Lab” (Florida, US). Venom from M. diastema was manually extracted from a single specimen collected in Los Tuxtlas, Veracruz, Mexico (Collection license # SGPS/DGVS/03459/15, SEMARNAT, Mexico) and kept at the “Herpetario Cantil” of IBt-UNAM, Cuernavaca, Mexico. The extracted venom was recovered using milli-Q H2O, centrifuged at 12,100 x g to remove cellular debris, lyophilized, and kept at 4 °C until use. Neurotoxins to be used as antigens for phage display campaigns were selected based on their abundance in the venoms of either M. fulvius6 or M. diastema, their high lethality in rodent models, and their similarity to toxins present in other North American coral snake venoms6,7,30,42,44,50,57,58. The two main PLA2 neurotoxins from M. fulvius venom (PLA2N and PLA2O), and the PLA2-containing fractions from Naja nigricollis venom (Nn19), Naja melanoleuca (Nm15), and Hemachatus haemachatus (Hh3), together with short-chain αNTx-containing fractions from M. diastema (DH), Naja haje (Nh1), Dendoaspis viridis (Dv1), and Hemachatus haemachatus (Hh1) were purified from whole venoms using reversed-phase high-performance liquid chromatography (RP-HPLC) with a C18 column using the gradient described previously6,7,55. In addition, α-cobratoxin from Naja kaouthia was purchased from Latoxan, and a PLA2 from the venom of Echis pyramidum was purified through size-exclusion chromatography employing a Superdex 75 Increase 10/300 GL column (Cytiva) pre-equilibrated with PBS. 5 mg/mL of venom diluted in PBS was added to the column and eluted at a flow rate of 0.5 mL/min in 500 µL fractions. Subsequently, these fractions underwent analysis via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a 10% Bis-Tris gel in MES buffer. Fractions displaying a band corresponding to the molecular weight of PLA2s (~14 kDa) were pooled for further use.
Expression and purification of recombinant toxins
Three short-chain αNTxs from the 3FTx family were used as target antigens for VHH discovery. Two of them, eurytoxin and αNTx DH, have previously been identified in coral snake venoms30,31, and the third one, scNTx, is a consensus protein, designed based on the sequences of eleven αNTxs from venoms of different elapids32. All three short-chain αNTxs were recombinantly produced in E. coli, using the SHuffle® T7 strain (New England Biolabs) for recombinant eurytoxin (rEury) and Origami Gold DE3 (Novagen®) for recombinant αNTx DH (rDH) and scNTx. Glycerol stocks of E. coli cells transformed with the pQE30 vector containing the toxins were kindly provided by Alejandro Olvera (rEury and rDH) and Dr Gerardo Corzo (scNTx) from IBt-UNAM. Cells from the glycerol stocks were used to inoculate 50 mL LB medium supplemented with ampicillin (80 µg/mL) and grown until an OD600 of 0.7 was reached. Next, 10 mL of these cultures were used to inoculate 1 L of LB medium and protein expression was induced by addition of 0.1 mM IPTG. Thereafter, the cells were cultured for 24 h at 16 °C and 250 rpm for protein expression. The toxins were purified from the supernatants by gravity flow purification using immobilized metal-ion affinity chromatography (HIS-Select® Nickel Affinity Gel, Merck Millipore). The slurry was equilibrated with PBS before adding the supernatant, after which the slurry was washed with PBS and the toxins eluted using 250 mM imidazole. The imidazole was removed by dialysis against PBS (Spectra/Por® dialysis membrane 3.5 kDa MWCO). Further purification was achieved using RP-HPLC on a C18 column equilibrated with 0.1% trifluoroacetic acid (TFA) and eluted using a gradient towards acetonitrile with 0.1% TFA7. All purified toxins were lyophilized and stored at 4 °C until use. The identity and integrity of the toxins were verified using mass spectrometry with an electrospray ionization system (ESI-MS) on an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific).
Biotinylation and mass spectrometry analysis of toxins
Lyophilized toxins were resuspended in PBS to a final concentration of approximately 2 mg/mL for biotinylation. The toxins were biotinylated using EZ-Link™ NHS-PEG4-Biotin (A39259, Thermo Fisher Scientific) at a 1:2 molar ratio (2 biotin for every toxin molecule) for 30 min at RT. Free biotin was removed using 2000 Da MWCO filter tubes (Sartorius). The toxin concentrations were determined using absorbance at 280 nm (NanoDrop, Thermo Fisher Scientific) and calculated based on their molar extinction coefficients, which were obtained in silico using the Expasy ProtParam tool (https://web.expasy.org/protparam/).
The molecular mass of all the toxins6,31 and the biotinylation ratio was determined by MALDI-TOF MS using an Ultraflex II TOF/TOF spectrometer (Bruker Daltonics).
Phage display selection campaigns and subcloning
To select for toxin-binding VHHs, phage display selection campaigns were performed using the VHH-displaying phage library17,19. In short, three consecutive rounds of selection were performed for three αNTxs (scNTx, rDH, and rEury) and two PLA2s (PLA2N and PLA2O), incubating the phage library with final toxin concentrations of 50 nM in the first two rounds and 10 nM in the third round. For toxins rDH and rEury, round 3 (round 3b) was repeated using 50 nM antigen due to the low enrichments observed. After the third round, the VHH-encoding genes were isolated from the glycerol stocks of the phage outputs, digested with PstI and Eco91I restriction endonucleases, subcloned into Xb-145 (a modified pHEN6 expression vector with an OmpA signal peptide and a C-terminal 3xFLAG and 6xHis-tag), and transformed into E. coli BL21 (DE3) cells33. Subsequently, individual VHH clones were picked into 500 µL of 2xYT medium supplemented with kanamycin (50 µg/mL) and glucose (2%) in 96-deep well plates and grown O/N at 30 °C and 800 rpm.
Screening of VHHs for antigen binding using DELFIA
For VHH expression, 10 µL of each overnight culture was used to inoculate 1 mL of autoinduction medium59 supplemented with kanamycin (50 µg/mL) in 96-deep-well plates, and the cultures were incubated O/N at 30 °C and 800 rpm. Thereafter, the plates were centrifuged for 10 min at 3000 x g, and the pellets were frozen at −20 °C O/N. The next day, the pellets were resuspended in 110 µL PBS, centrifuged for 10 min at 4500 x g, and the supernatants (the periplasmic fractions in which the majority of the VHH products were expected to exist) were transferred to a 96-well plate and stored at −20 °C until use. The VHHs were screened for binding using an expression-normalized capture DELFIA18, where a 1:100 dilution of VHH-containing periplasmic fractions in 3% milk-PBS was added to the 96-well MaxiSorp plates (Thermo Fisher Scientific) coated with 2.5 µg/mL of anti-FLAG M2 antibody O/N (F3165, Sigma-Aldrich), followed by addition of 100 nM of biotinylated toxins rEury, rDH, or scNTx. The bound toxins were detected with 0.2 ng/µL of Europium-labelled streptavidin diluted in DELFIA assay buffer (Perkin Elmer), followed by addition of 100 µL DELFIA enhancement solution (Perkin Elmer) per well. Binding was assessed via measuring Time-Resolved Fluorescence (TRF) signal at 337 nm (excitation) and 615 nm (emission), using a plate reader (VICTOR® Nivo™, PerkinElmer).
The plasmids encoding VHH binders with the highest signal against each target antigen were purified (GeneJET Plasmid MiniPrep kit, Thermo Fisher Scientific) according to the manufacturer’s protocol and sequenced using the M13rev-29 primer (Eurofin Genomics). The VHH frameworks and CDRs were annotated and analyzed to identify unique clones (CLC Main workbench v22.0.2).
Expression and purification of VHHs
In total, six VHHs were selected for expression and purification: three from the PLA2 selection campaigns (TPL0637_01_A01, TPL0637_01_A07, and TPL0638_01_C09) and three from the αNTx selection campaigns (TPL0629_01_D11, TPL0629_01_A07 and TPL0629_01_G06). VHH expression was performed using E. coli BL21 (DE3) in TB medium supplemented with kanamycin (50 µg/mL), glucose (0.1% w/v), and MgSO4 (1 mM). The cells were grown at 37 °C and 220 rpm until OD600 reached 0.5. Subsequently, 0.5 mM IPTG was added to induce protein expression for 16 h at 30 °C and 220 rpm. The cells were collected by centrifugation at 4,000 x g for 15 min at 4 °C and stored at −20 °C. The frozen cells were then resuspended in cold PBS supplemented with 10 mM imidazole and an EDTA-free protease inhibitor cocktail (Roche). Next, the periplasmic fractions containing the His-tagged VHHs were collected by centrifugation at 20,000 x g for 45 min at 4 °C. The VHHs were then captured on an affinity resin by gravity flow (HIS-Select® Nickel Affinity Gel, Merck Millipore). Unbound proteins were washed away with wash buffer (PBS with 200 mM NaCl and 20 mM imidazole). The VHHs were then eluted from the resin (PBS with 200 mM NaCl and 250 mM imidazole), after which the imidazole was removed by dialyzing against PBS (SnakeSkinTM dialysis tubing, Thermo Fisher Scientific; 3.5 kDa MWCO). The purity of the VHHs was analyzed by SDS-PAGE and the VHH concentration was determined by absorbance at 280 nm (NanoDrop, Thermo Fisher Scientific), and calculated by their molar extinction coefficients which were determined using the Expasy ProtParam Tool (https://web.expasy.org/protparam/).
Binding analysis in dose-response DELFIA
To assess cross-reactivity of the VHHs, dose-response DELFIAs were performed as described for the screening of VHHs with a few exceptions. These included adding 5 µg/mL of purified VHHs instead of VHH-containing periplasmic fractions to the anti-FLAG coated plates. Also, instead of using a single toxin concentration, the targets were first diluted to 1 µM, and then titrated 1:3 in 10 consecutive dilution steps and added to the plate. All other steps were identical to those described earlier. The targets consisted of a purified PLA2 from M. fulvius venom (PLA2N), PLA2-containing fractions from the venom of H. haemachatus (Hh3), N. melanoleuca (Nm15), N. nigricollis (Nn19), and E. pyramidum, a purified αNTx from the venom of M. diastema (αNTx DH), αNTx-containing fractions from the venoms of N. haje (Nh1), D. viridis (Dv1), and H. haemachatus (Hh1), and α-cobratoxin purchased from Latoxan.
Binding analysis using bio-layer interferometry
Binding kinetics between the purified VHHs and their specific toxin targets were analyzed using bio-layer interferometry. Measurements were performed in kinetics buffer (PBS and 0.02% Tween 20; ForteBio) at 30 °C using an Octet RED 96 instrument (ForteBio). Biotinylated PLA2N or αNTx DH with a final concentration of 1 µg/mL were loaded onto streptavidin biosensors (Sartorius) until a thickness of approximately 0.9 nm was reached. Toxin-loaded biosensors were dipped into five VHH concentrations (200, 66.7, 22.2, 7.4, 2.5, and 0.8 nM). The association of VHHs to the toxins was measured for 600 s, followed by the dissociation for 600 s by incubating the biosensors in kinetics buffer. Sensors were regenerated by two rounds of 5 s incubations in Glycine at pH 1.5, followed by kinetics buffer before measuring the binding kinetics of the next VHH-toxin pair. Two reference measurements, one without biotinylated toxin and the highest concentration of VHH, and one with biotinylated toxin but without any VHH, were subtracted from all curves. All data were analyzed using Octet® Analysis Studio 12.2.2.26 (ForteBio).
In vitro neutralization of enzymatic PLA2 activity
Determination of PLA2 enzymatic activity was performed using the fluorometric EnzChek™ Phospholipase A2 Assay Kit (Invitrogen) according to manufacturer’s protocol. Fluorescence was measured using a plate reader (VICTOR® Nivo™, PerkinElmer) at an excitation wavelength of 480 nm and an emission wavelength of 530 nm. Measurements were made immediately after substrate addition and then every 30 s for 10 min to verify the linearity of the kinetics. The enzymatic activity was defined as the relative fluorescence obtained 5 min after substrate addition.
Neutralization of enzymatic activity was assessed by incubating 0.1 mg/mL of purified PLA2s or PLA2-containing fractions from different elapid snake venoms (PLA2N from the venom of M. fulvius, Nn19 from Naja nigricollis, and Hh3 from H, haemachatus) with a 1:20 toxin to VHH molar ratio of each anti-PLA2 VHH for 30 min at RT. PLA2 activity was determined for each of the mixtures in duplicate. The PLA2 activity in the presence of the VHHs was normalized by setting the activity of the toxin incubated with buffer only to 100%.
In vitro neutralization of αNTx mediated blocking of nAChR activity (Automated Patch Clamp electrophysiology)
Automated planar whole-cell patch clamp experiments were conducted to evaluate the neutralizing capacity of the discovered VHHs on αNTx DH and scNTx-mediated blocking of nAChR activity. All electrophysiology experiments were performed using a human-derived Rhabdomyosarcoma RD cell line (American Type Culture Collection, ATCC), which endogenously expresses muscle-type nAChRs (α1, β1, δ, and γ-subunit), on a Qube 384 automated patch clamp platform (Sophion Bioscience) with 384-channel patch chips (patch hole resistance 2.00 ± 0.02 MΩ) as described elsewhere19. The nAChR-mediated currents were elicited by 70 µM acetylcholine (ACh) (corresponding to approximately the EC80 concentration). A second ACh addition was used to evaluate the toxin effect in combination with each of the three discovered anti αNTx VHHs at different concentrations (5, 15, 45, and 135 nM). The toxin concentration (αNTx DH = 15 nM; scNTx = 5 nM) was chosen to be approximately the previously determined IC80 value, which is the toxin concentration that inhibits 80% of the maximum ACh current. Toxins and VHHs were preincubated for at least 30 min before addition to the cells and the patched cells were incubated with the toxin-VHH mixtures for 5 min before the second ACh addition. The inhibitory effect of the toxins on the elicited ACh current was normalized to the full ACh response and averaged in the group (n = 8). The data was analyzed with Sophion Analyzer v6.6.70 (Sophion Bioscience) and GraphPad Prism v10.
Design, expression, and purification of bivalent-VHH and VHH-Fc constructs
For expression of bivalent TPL0629_01_D11, the pUC57 vector containing VHH-(GGGGS)3-VHH was purchased as a synthetic gene from GenScript. The plasmid was transformed into XL1-Blue cells (Agilent), amplified, and purified using a miniprep kit following the manufacturer’s instruction (GeneJET Plasmid MiniPrep kit, Thermo Fisher Scientific). Afterwards, the purified plasmid was digested using NotI and PstI restriction enzymes (New England Biolabs) and ligated into the Xb-145 expression vector using T4 DNA ligase (New England Biolabs). After transformation into BL21(DE3) cells, a positive transformant was used for the expression and purification of a bivalent TPL0629_01_D11 construct similarly as explained for monovalent VHHs.
For expression of VHH-Fc, the nucleotide sequence of the constant heavy chain domain 2 and 3 from an human IgG1 antibody, harboring the LALA/YTE60,61 mutations, was PCR amplified from the proprietary pINT319 vector and subjected to EcoRI and NotI digestion. Thereafter, the constant heavy chain domain 1 from the proprietary plasmid pINT1219 was excised using the EcoRI and NotI restriction enzymes and replaced with the PCR amplified constant heavy chain domain 2 and 3.
The nucleotide sequence of the TPL0629_01_D11 VHH was PCR amplified and integrated into the newly constructed vector using NEBuilder assembly, resulting in the generation of the expression plasmid TPL0629_01_D11-Fc (LALA, YTE). ExpiCHO cells (Thermo Fisher Scientific) were cultured and transfected according to the manufacturer’s protocol using the plasmid at a concentration of 1 µg DNA/mL and ExpiFectamine. Transiently transfected cells were cultivated for four days at 125 rpm, 37 °C, 8% CO2, and 70% humidity. Following incubation, cells were harvested, and VHH-Fc in the supernatant was purified using affinity chromatography on a MabSelect SuRe column (Neo Biotech).
In vivo neutralization experiments
All in vivo experiments were performed with groups of three CD1 mice between 18 and 20 g of total body weight and indistinct sex. All mice were provided by the animal facility of IBt-UNAM and were kept under 12 h light and dark cycles with food and water ad libitum, ambient temperature of 18–24 °C and approximately 60% relative humidity. For neutralization of whole coral snake venoms, two coral snake species (i.e., M. fulvius and M. diastema), whose venoms contain identical or very similar toxins to the ones used as antigens in the phage display selection campaigns, were chosen. Toxin to VHH molar ratios were calculated based on the approximate abundance of PLA2s and 3FTxs in the venoms, obtained from proteomic data6. The number of 3FTx or PLA2 molecules present in 3 LD50s of venom was then used to calculate the necessary amount of VHHs in the mixture (Supplementary Table 4).
Determination of LD50s for purified toxins and whole venoms
LD50s were determined for PLA2N (M. fulvius) and αNTx DH (M. diastema) toxins and the whole venoms of M. fulvius and M. diastema using the i.v. and s.c. routes. Groups of three mice were injected with varying doses of the toxin or whole venom in a final volume of 500 µL PBS for i.v. and 100 µL PBS for s.c. injection. The survival percentage was determined 24 h after injection, and the data was analyzed using a non-linear regression (semi-logarithmic dose-response curve)62.
Preincubation experiments
For preincubation experiments, 3 LD50s of each toxin or venom were combined with their corresponding VHH or VHH mixture using a range of toxin to VHH molar ratios going from 1:1 to 1:10 in a total volume of 500 µL of PBS (Supplementary Tables 2 and 5), following the guidelines of the Mexican Pharmacopeia (9th Edition)63. Due to a revision in the ethical guidelines and protocols, the injection volume was decreased to 250 µL during the whole venom neutralization experiments. For comparison, the commercial polyclonal antivenom, Coralmyn, which is composed of purified equine F(ab’)2 fragments and is currently the only treatment available in Mexico for coral snake envenomation, was used. The antivenom was combined with either the venom of M. fulvius or M. diastema at an approximate venom to antivenom molar ratio of 1:5 (Supplementary Table 5). One vial of Coralmyn (Batch # B-2H-12) was resuspended in 1 mL of injectable saline (provided by the manufacturer) and protein concentration was determined by measuring absorbance at 280 nm and corrected using an estimated extinction coefficient for F(ab’)2 of 1.44. To calculate venom to Coralmyn molar ratio, 100% of the protein content of Coralmyn was assumed to be F(ab)’2. The blends were preincubated at 37 °C for 30 min and injected into groups of 3 mice using the i.v. route. The mice were observed during the first 3 h and then approximately every 6 h for appearance of envenomation signs. The percentage of survival was calculated up to 24 h after the injection.
Rescue experiments
Rescue experiments were designed to better represent real envenomation, where the toxin is injected first and then the therapeutic molecule is administered using the i.v. route. In these experiments, mice were envenomed using the s.c. route with 3 LD50s of each toxin in a final volume of 100 µL PBS. Immediately after toxin injection, the corresponding VHH or VHH construct was injected using the i.v. route in a total volume of 500 µL PBS (Supplementary Table 3). The experiments were performed using a range of toxin to VHH molar ratios going from 1:1 to 1:10. Mice were observed during the first 3 h and then approximately every 6 h for the appearance of envenomation signs. The percentage of survival was calculated up to 24 h after the injection.
Statistical analysis
To estimate the significance of the results obtained in the in vivo experiments, we performed a Mantel-Cox log-rank test64. Data were compared either to the negative control (PBS only) or to the commercial antivenom, Coralmyn. The significance value was set to α = 0.05, and therefore P-values higher than this were considered as non-significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the data supporting the present manuscript is available in the form of Source Data Files and in the supplementary material. Relevant VHH and toxin sequences as well as detailed information on in vivo experiments are provided in the Supplementary Material. Raw data and analyses performed for the figures are available as Source Data Files. Source data are provided with this paper.
References
Laustsen, A. H. How can monoclonal antibodies be harnessed against neglected tropical diseases and other infectious diseases? Expert Opin. Drug Discov. 14, 1103–1112 (2019).
Burton, D. R., Poignard, P., Stanfield, R. L. & Wilson, I. A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337, 183–186 (2012).
Julg, B. et al. Safety and antiviral activity of triple combination broadly neutralizing monoclonal antibody therapy against HIV-1: a phase 1 clinical trial. Nat. Med. 28, 1288–1296 (2022).
Chippaux, J.-P. Incidence and mortality due to snakebite in the Americas. PLoS Negl. Trop. Dis. 11, e0005662 (2017).
Lomonte, B. et al. Venoms of Micrurus coral snakes: Evolutionary trends in compositional patterns emerging from proteomic analyses. Toxicon 122, 7–25 (2016).
Vergara, I. et al. Eastern coral snake Micrurus fulvius venom toxicity in mice is mainly determined by neurotoxic phospholipases A2. J. Proteom. 105, 295–306 (2014).
Bénard-Valle, M. et al. Functional, proteomic and transcriptomic characterization of the venom from Micrurus browni browni: Identification of the first lethal multimeric neurotoxin in coral snake venom. J. Proteom. 225, 103863 (2020).
Bucaretchi, F. et al. Coral snake bites (Micrurus spp.) in Brazil: a review of literature reports. Clin. Toxicol. 54, 222–234 (2016).
De Roodt, A. R. et al. Effectiveness of two common antivenoms for North, Central, and South American Micrurus envenomations. J. Toxicol. - Clin. Toxicol. 42, 171–178 (2004).
Gutiérrez, J. M. Snakebite Envenoming: A Public Health Perspective. in Public Health – Methodology, Environmental and Systems Issues (ed. Maddock, P. J.) (InTech). https://doi.org/10.5772/36076 (2012).
Sanchez, E. E., Lopez-Johnston, J. C., Rodrıguez-Acosta, A. & Perez, J. C. Neutralization of two North American coral snake venoms with United States and Mexican antivenoms. Toxicon 51, 297–303 (2008).
Laustsen, A. H. et al. Pros and cons of different therapeutic antibody formats for recombinant antivenom development. Toxicon 146, 151–175 (2018).
Kini, R. M., Sidhu, S. S. & Laustsen, A. H. Biosynthetic oligoclonal antivenom (BOA) for snakebite and next-generation treatments for snakebite victims. Toxins 10, 534 (2018).
Archundia, I. G. et al. Assessment of neutralization of Micrurus venoms with a blend of anti-Micrurus tener and anti-ScNtx antibodies. Vaccine 39, 1000–1006 (2021).
Laustsen, A. H. et al. Exploration of immunoglobulin transcriptomes from mice immunized with three-finger toxins and phospholipases A2 from the Central American coral snake, Micrurus nigrocinctus. PeerJ 5, e2924 (2017).
Laustsen, A. H., Greiff, V., Karatt-Vellatt, A., Muyldermans, S. & Jenkins, T. P. Animal immunization, in vitro display technologies, and machine learning for antibody discovery. Trends Biotechnol. 39, 1263–1273 (2021).
Ahmadi, S. et al. An in vitro methodology for discovering broadly-neutralizing monoclonal antibodies. Nat. Sci. Rep. 10, 10765 (2020).
Laustsen, A. H. et al. In vivo neutralization of dendrotoxin-mediated neurotoxicity of black mamba venom by oligoclonal human IgG antibodies. Nat. Commun. 9, 1–9 (2018).
Ledsgaard, L. et al. Discovery and optimization of a broadly-neutralizing human monoclonal antibody against long-chain α-neurotoxins from snakes. Nat. Commun. 14, 682 (2023).
Laustsen, A. H., Lohse, B., Lomonte, B., Engmark, M. & Gutiérrez, J. M. Selecting key toxins for focused development of elapid snake antivenoms and inhibitors guided by a Toxicity Score. Toxicon 104, 43–45 (2015).
Sørensen, C. V. et al. Cross-reactivity trends when selecting scFv antibodies against snake toxins using a phage display-based cross-panning strategy. Sci. Rep. 13, 1–10 (2023).
Rivera‐de‐Torre, E. et al. Discovery of broadly‐neutralizing antibodies against brown recluse spider and Gadim scorpion sphingomyelinases using consensus toxins as antigens. Protein Sci. 33, 1–15 (2024).
Laustsen, A. H. Antivenom in the Age of Recombinant DNA Technology. in Handbook of Venoms and Toxins of Reptiles (ed. Mackessy, S. P.) 499–510 (CRC Press. Taylor & Francis Group). https://doi.org/10.1201/9780429054204-38 (2021).
Pucca, M. B. et al. History of envenoming therapy and current perspectives. Front. Immunol. 10, 1–13 (2019).
Jenkins, T. P. & Laustsen, A. H. Cost of manufacturing for recombinant snakebite antivenoms. Front. Bioeng. Biotechnol. 8, 1–13 (2020).
Jenkins, T. P. et al. Toxin neutralization using alternative binding proteins. Toxins 11, 1–28 (2019).
Dumoulin, M. et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 11, 500–515 (2002).
Hmila, I. et al. VHH, bivalent domains and chimeric heavy chain-only antibodies with high neutralizing efficacy for scorpion toxin AahI′. Mol. Immunol. 45, 3847–3856 (2008).
Wade, J. et al. Generation of multivalent nanobody-based proteins with improved neutralization of long α-neurotoxins from elapid snakes. Bioconjug. Chem. 33, 1494–1504 (2022).
Bénard-Valle, M. et al. Protein composition and biochemical characterization of venom from Sonoran Coral Snakes (Micruroides euryxanthus). Biochimie 182, 206–216 (2021).
Guerrero-Garzón, J. F. et al. Cloning and sequencing of three-finger toxins from the venom glands of four Micrurus species from Mexico and heterologous expression of an alpha-neurotoxin from Micrurus diastema. Biochimie 147, 114–121 (2018).
De La Rosa, G., Corrales-García, L. L., Rodriguez-Ruiz, X. Estuardo López-Vera. & Corzo, G. Short-chain consensus alpha-neurotoxin: a synthetic 60-mer peptide with generic traits and enhanced immunogenic properties. Amino Acids 50, 885–895 (2018).
Wouters, Y., Jaspers, T., De Strooper, B. & Dewilde, M. Identification and in vivo characterization of a brain-penetrating nanobody. Fluids Barriers CNS 17, 4–13 (2020).
Montagut, C. et al. Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti-EGFR therapy and molecularly selected by circulating tumor DNA analyses a phase 2 randomized clinical trial. JAMA Oncol. 4, 1–9 (2018).
PREVAIL II Writing Group A Randomized, Controlled Trial of ZMapp for Ebola Virus Infection. N. Engl. J. Med. 375, 1448–1456 (2016).
O’Brien, M. P. et al. Effect of subcutaneous Casirivimab and Imdevimab antibody combination vs placebo on development of symptomatic COVID-19 in early asymptomatic SARS-CoV-2 infection: A randomized clinical trial. Jama 327, 432–441 (2022).
Casewell, N. R., Jackson, T. N. W., Laustsen, A. H. & Sunagar, K. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 41, 570–581 (2020).
Rey-Suárez, P., Núñez, V., Fernández, J. & Lomonte, B. Integrative characterization of the venom of the coral snake Micrurus dumerilii (Elapidae) from Colombia: Proteome, toxicity, and cross-neutralization by antivenom. J. Proteom. 136, 262–273 (2016).
Ledsgaard, L. et al. In vitro discovery of a human monoclonal antibody that neutralizes lethality of cobra snake venom. MAbs 14, 1–11 (2022).
Miersch, S. et al. Synthetic antibodies block receptor binding and current-inhibiting effects of α-cobratoxin from Naja kaouthia. Protein Sci. 31, e4296 (2022).
Laustsen, A. H. Recombinant snake antivenoms get closer to the clinic. Trends Immunol. 45, 225–227 (2024).
Bénard-Valle, M., Carbajal-Saucedo, A., De Roodt, A., López-Vera, E. & Alagón, A. Biochemical characterization of the venom of the coral snake Micrurus tener and comparative biological activities in the mouse and a reptile model. Toxicon 77, 6–15 (2014).
Rey-Suárez, P. et al. Mipartoxin-I, a novel three-finger toxin, is the major neurotoxic component in the venom of the redtail coral snake Micrurus mipartitus (Elapidae). Toxicon 60, 851–863 (2012).
Aird, S. D. et al. Coralsnake venomics: Analyses of venom gland transcriptomes and proteomes of six Brazilian taxa. Toxins 9, 187 (2017).
Gutiérrez, J. M. et al. Preclinical evaluation of the efficacy of antivenoms for snakebite envenoming: State-of-the-art and challenges ahead. Toxins 9, 1–22 (2017).
Neri-castro, E. et al. Neotropical rattlesnake (Crotalus simus) pharmacokinetics in lymph and blood using an ovine model. Toxins 12, 1–24 (2020).
Yap, M. K. K., Tan, N. H., Sim, S. M. & Fung, S. Y. Toxicokinetics of Naja sputatrix (Javan spitting cobra) venom following intramuscular and intravenous administrations of the venom into rabbits. Toxicon 68, 18–23 (2013).
Paniagua, D. et al. Lymphatic route of transport and pharmacokinetics of Micrurus fulvius (Coral snake) venom in sheep. Lymphology 45, 144–153 (2012).
Fernández, J. et al. Snake venomics of Micrurus alleni and Micrurus mosquitensis from the Caribbean region of Costa Rica reveals two divergent compositional patterns in New World elapids. Toxicon 107, 217–233 (2015).
Fernández, J. et al. Venomic and antivenomic analyses of the Central American coral snake, Micrurus nigrocinctus (Elapidae). J. Proteome Res. 10, 1816–1827 (2011).
Yin, M. et al. Evolution of nanobodies specific for BCL11A. Proc. Natl Acad. Sci. 120, e2218959120(2023).
Khan, A. et al. Toward real-world automated antibody design with combinatorial Bayesian optimization. Cell Rep. Methods 3, 100374 (2023).
Gutierrez, J. M., León, G. & Lomonte, B. Pharmacokinetic-Pharmacodynamic relationships of immunoglobulin therapy for envenomation. Clin. Pharmacokinet. 42, 721–741 (2003).
European Parliament Directive 2010/63/EU - On the protection of animals used for scientific purposes. J. Eur. Union 276, 33–79 (2010).
Nguyen, G. T. T. et al. High-throughput proteomics and in vitro functional characterization of the 26 medically most important elapids and vipers from sub-Saharan Africa. GigaScience 11, 1–15 (2022).
Pardon, E. et al. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 9, 674–693 (2014).
Lomonte, B., Sasa, M., Rey-Suárez, P., Bryan, W. & Gutiérrez, J. M. Venom of the coral snake Micrurus clarki: Proteomic profile, toxicity, immunological cross-neutralization, and characterization of a three-finger toxin. Toxins 8, 138 (2016).
Margres, M. J., Aronow, K., Loyacano, J. & Rokyta, D. R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics 14, 531 (2013).
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Dall’Acqua, W. F. et al. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: Biological consequences. J. Immunol. 169, 5171–5180 (2002).
Xu, D. et al. In vitro characterization of five humanized OKT3 effector function variant antibodies. Cell. Immunol. 200, 16–26 (2000).
Casasola, A. et al. Paraspecific neutralization of the venom of African species of cobra by an equine antiserum against Naja melanoleuca: A comparative study. Toxicon 53, 602–608 (2009).
Secretaria de Salud. Métodos de productos biológicos. in Farmacopea de los Estados Unidos Mexicanos. Vol II 2181 (Comisión Permanente de la Farmacopea de los Estados Unidos Mexicanos (FEUM), 2008).
Bewick, V., Cheek, L. & Ball, J. Statistics review 12: Survival analysis. Crit. Care 8, 389–394 (2004).
Acknowledgements
The VHH expression vector was a kind gift from the laboratory of Professor Bart De Strooper at the KU Leuven and VIB. The authors sincerely thank Alejandro Olvera and Dr. Gerardo Corzo for kindly providing the plasmids for expression of the αNTxs and Manuel Yañez Mendoza for his assistance during these expressions. We also thank Dr. Edgar Neri-Castro and Vanesa Gómez-Zarzosa for their assistance in coral snake handling and venom extraction. The authors also thank Jack Facente for kindly providing the pool of M. fulvius venom used for the present research as well as Lorenzo Seneci and Mathieu Roumet for proofreading of the present manuscript. Finally, the authors thank Sergio González and Dr. Elizabeth Mata from the animal facility at IBt, UNAM, for mice reproduction and wellbeing management, as well as training regarding good practices for in vivo experiments. AHL is supported by a grant from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program [850974], a grant from the Villum Foundation [00025302], and a grant from Wellcome [221702/Z/20/Z]. TPJ has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement [713683] (COFUNDfellowsDTU). MBV is supported by a Eurotech Postdoctoral fellowship from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement [899987]. AA has received funding from the Mexican Consejo Nacional de Ciencia y Tecnología, FORDECyT-PRONAII [303045]. SS, SPB, and BGV are supported by a grant from the Novo Nordisk Foundation [NNF20SA0066621].
Author information
Authors and Affiliations
Contributions
Conceptualization: M.B.V., A.H.L. Methodology: M.B.V., A.L., Y.W., G.T.N., T.W.E., A.G.M., G.R.B., K.B., T.P.J., S.A., S.S., S.P.B., A.H.L. Investigation: M.B.V., A.L., Y.W., G.T.N., T.W.E., F.J., H.E., A.G.M., G.R.B., K.B., S.A., C.H.D., S.S., S.P.B. Visualization: M.B.V., A.L., Y.W. Funding acquisition: A.A., M.B.V., T.P.J., B.G.V., A.H.L. Project administration: A.L., M.B.V., A.H.L. Resources: A.A., A.H.L. Supervision: A.L., A.H.L. Writing – original draft: M.B.V., A.L., Y.W., T.P.J. Writing – review & editing: M.B.V., A.L., A.H.L., T.P.J., A.A., S.A., A.H.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: M.B.V., A.L., and A.H.L. are inventors on a submitted patent application (EP23192644.5), owned by the Technical University of Denmark. The remaining authors declare that they have no competing interests.
Peer review
Peer review information
Nature Communications thanks Kartik Sunagar and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Benard-Valle, M., Wouters, Y., Ljungars, A. et al. In vivo neutralization of coral snake venoms with an oligoclonal nanobody mixture in a murine challenge model. Nat Commun 15, 4310 (2024). https://doi.org/10.1038/s41467-024-48539-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-48539-z
- Springer Nature Limited