Abstract
SETD3 is a member of SET-domain containing methyltransferase family, which plays critical roles in various biological events. It has been shown that SETD3 could regulate the transcription of myogenic regulatory genes in C2C12 differentiation and promote myoblast determination. However, how SETD3 is regulated during myoblast differentiation is still unknown. Here, we report that two important microRNAs (miRNAs) could repress SETD3 and negatively contribute to myoblast differentiation. Using microRNA (miRNA) prediction engines, we identify and characterize miR-15b and miR-322 as the primary miRNAs that repress the expression of SETD3 through directly targeting the 3’-untranslated region of SETD3 gene. Functionally, overexpression of miR-15b or miR-322 leads to the repression of endogenous SETD3 expression and the inhibition of myoblast differentiation, whereas inhibition of miR-15b or miR-322 derepresses endogenous SETD3 expression and facilitates myoblast differentiation. In addition, knockdown SETD3 in miR-15b or miR-322 repressed myoblasts is able to rescue the facilitated differentiation phenotype. More interestingly, we revealed that transcription factor E2F1 or FAM3B positively or negatively regulates miR-15b or miR-322 expression, respectively, during muscle cell differentiation, which in turn affects SETD3 expression. Therefore, our results establish two parallel cascade regulatory pathways, in which transcription factors regulate microRNAs fates, thereby controlling SETD3 expression and eventually determining skeletal muscle differentiation.
Similar content being viewed by others
Introduction
Skeletal muscle differentiation is a complex process orchestrated by a family of myogenic regulatory factors (MRFs), including MyoD, myogenin, MRF4, and Myf51,2. Expression of MyoD and Myf5 in the initial stages of differentiation induces expression of myogenin and muscle-specific transcription factors MEF2, whereas myogenin and MRF4 are expressed in the late stages of differentiation to activate the myogenic program by induction of muscle gene expression and silence of cell cycle-related gene expression2,3,4. Moreover, the functional interplay between key myogenic transcriptional factors and additional regulators is also critical for determining muscle cell fate and myotube/myofibers formation2,5,6.
MicroRNAs (miRNAs) modulate gene expression at the post-transcriptional level either by promoting mRNA degradation or inhibiting translation through complementary targeting 3’ untranslated regions (3’-UTRs) of specific mRNAs2,6. Many studies have demonstrated that miRNAs participate in skeletal muscle differentiation. The muscle-specific miRNAs, miR-206, miR-1, and miR-133, are abundantly expressed during skeletal muscle differentiation, and promote muscle differentiation by inhibition specific transcription repressors7,8,9,10. In addition, many non-muscle specific miRNAs also regulate muscle differentiation by post-transcriptional mechanisms that affect the presence and functions of the myogenic factors, either positively or negatively.
Our previous work focused on studying the biological roles of SETD3, which has been reported as a histone H3 Lys4 and Lys36 methyltransferase11. But very recent two studies clearly demonstrated that SETD3 is an actin-specific histidine methyltransferase12,13. We have shown that SETD3 is a cell-cycle regulated protein, and abnormal high level of SETD3 would lead to liver tumorigenesis14. A previous study has suggested that SETD3 is capable to interacting with MyoD and synergistically binding to the promoter of several muscle-related genes, thereby promoting muscle cell differentiation11. Knockdown of SETD3 markedly impairs the differentiation processes, indicating its important role in muscle differentiation. However, how SETD3 is regulated during this process is completely unknown.
In this study, we hypothesized that SETD3 gene is post-transcriptionally repressed by miRNAs. We uncovered that miR-15b and miR-322 could repress SETD3 expression by targeting the 3′-UTR region in skeletal muscle cells. Furthermore, we revealed that two known transcription factors, E2F1 and FAM3B, could regulate miR-15b or miR-322 expression, respectively, during muscle cell differentiation. Thus, our results established a regulatory network between transcription factors, miRNAs, and an epigenetic modifier SETD3, which highlights a protein-microRNA involved cascade regulatory mechanism during skeletal muscle differentiation.
Results
SETD3 is required for C2C12 cell differentiation
Previous study suggested that SETD3 regulates muscle differentiation11. To confirm this, we first generated a monoclonal SETD3 antibody to detect endogenous SETD3 protein. This anti-SETD3 antibody specifically recognizes the SETD3 protein, as detected SETD3 signal was diminished when SETD3 gene was knocked out in Hela S3 cells and overexpression of SETD3 constructs from either human or mouse species in the SETD3 knockout cell line displayed specific bands (supplementary Fig. S1a). In addition, this anti-SETD3 antibody also recognizes endogenous SETD3 in C2C12 mouse myoblast cells, and knockdown of mouse SETD3 by stable expression of two different shSETD3 constructs exhibited significant reduction of SETD3 level, indicating its specificity and species reactivity against mouse homolog SETD3 as well (supplementary Fig. S1a). Next, to examine whether SETD3 is required for cell differentiation, C2C12 cells was induced by cultured in the differentiation medium (DM), and expression of SETD3 in both transcriptional levels and protein levels were examined. Consistent with previous results, transcription levels of several key regulatory factors including MYF5, MYOG, TNNT2/Troponin, and MYH1/MYHC were gradually increased during differentiation, with a similar trend of SETD3 expression, indicating cell differentiation occurred (Fig. S1b)2. Intriguingly, we found that the protein levels of SETD3 displayed an increase at the early stage of differentiation, but showed a reduction when MHC protein was significantly accumulated, which may suggest a complicated regulatory mechanism of SETD3 involved in muscle differentiation (Fig. S1c). To rule out the possibility that the reduction of SETD3 protein level at the late stage of differentiation is due to our home-made antibody recognition issue, a commercial available antibody was utilized to examine SETD3 protein levels, and a similar expression pattern of SETD3 protein was observed (Fig. S1c). In addition, both antibodies were verified using two different synthesized siRNA oligos targeting SETD3, which confirmed the specificity of both antibodies (Fig. S1d). Consistent with previous report that knockdown of SETD3 severely slows muscle cell differentiation based on the observation of cell morphology and differentiation gene expression11, we also observed knockdown of SETD3 remarkably delayed cell differentiation, based on the divergence of cell morphology (Fig. S1e, f). Moreover, the protein levels of MHC as well as the mRNA levels of various differentiation markers were significantly reduced compared to the control cells during the progression (Fig. S1g, h). Therefore, our data support that SETD3 is required for C2C12 muscle cell differentiation.
Identification of miRNAs that might affect SETD3 expression
We are interested in how SETD3 levels are regulated at post-transcriptional levels during cell differentiation. Thus, we attempted to identify whether miRNAs might regulate expression of SETD3. To this end, the 3′ end of untranslated region (3′-UTR, nt 1786-2541) of mouse SETD3 gene was selected for searching potential miRNAs using miRanda and TargetScan softwares15,16. Based on the predicted scores, we obtained several potential miRNAs and the top 5 candidates were selected (Fig. 1a). Interestingly, the binding regions of these five potential candidates are nearly identical, which are located from nt 1872 to nt 1894 in the 3′-UTR of SETD3 gene. To identify which miRNAs might regulate SETD3 expression, we first cloned the full-length (756 nt) 3′-UTR of the mouse SETD3 gene and inserted into the downstream of a dual-luciferase reporter construct17. After the reporter construct was transfected into 293 T cells, we observed that only miR-15b or miR-322, but not other tested miRNAs, inhibited luciferase activity compared with the control construct (Fig. 1b). MiR-410 has been known to be not involved in regulation of SETD3 expression, which served as a negative control. To further confirm this, a short 3′-UTR sequence (23 nt) that only contains the predicted binding sites shared by all the 5 miRNAs were inserted into the downstream of a dual-luciferase reporter construct. Again, we found that only miR-15b and miR-322 showed repressive effect towards luciferase activity (Fig. 1c). Of note, miR-15b and miR-322 share the same 3′-UTR region of SETD3 gene, but with a slight difference in the seed region (Fig. 1d). Moreover, these two miRNAs are highly conserved among different species, suggesting their intrinsic function (Fig. 1e).
a List the top 5 of predicted miRNAs targeting the 3′-UTR of mouse SETD3 gene. Score represents the potential capability of miRNA binding to 3′-UTR of SETD3 gene. b, c Dual-luciferase reporter assays were performed using the indicated miRNAs co-transfected with a dual-luciferase reporter construct containing either a long 3′-UTR fragment (756 bp, panel b) or a short 3′-UTR fragment (23 bp, panel c) of SETD3 gene. Dual-luciferase activities were normalized to the mock control, which was set as 1. Experiments were performed in three biological duplicates, and data are presented as mean ± SD. d Sequence alignment of mouse miR-15b or miR-322 with the 3′-UTR of SETD3 gene was shown. e Homologous comparisons of miR-15b or miR-322 in different species. mmu Mus musculus, hsa Homo sapiens, rno Rattus norvegicus
MiR-15b and miR-322 directly targeted the 3′-UTR of SETD3 gene
To validate whether miR-15b and miR-322 indeed targeted the 3′-UTR of SETD3 gene, the predicted seed sequences of the short 3′-UTR in the luciferase reporter were mutated, and luciferase assays were performed as described above (Fig. 2a). We noticed that, when the reporter construct containing triple repeats of the short 3′-UTR of SETD3, the luciferase activity was repressed by both miR-15b and miR-322 more efficiently than the one containing a single copy of the short 3′-UTR of SETD3. In contrast, the luciferase activity remained invariable after the seed sequences in the 3′-UTR were mutated, compared to the control sample (Fig. 2b, c). When the indicated luciferase reporter constructs were transfected into cells that stably express pri-miR-15b or pri-miR-322, the luciferase activities were repressed by these miRNAs, but not by the empty luciferase reporter vector (Fig. 2d, e). In contrast, cotransfection of the indicated miRNA inhibitors with the luciferase reporter constructs into cells, the luciferase activities were enhanced compared to the control, suggesting their repressive roles of miRNAs in SETD3 gene expression (Fig. 2f). Therefore, these results provided clear evidence showing that miR-15b or miR-322 can directly target the 3′-UTR of SETD3 gene in vitro.
a Sequence alignment of the short 3′-UTR fragment of the wild-type or the mutant SETD3 was shown. The mutated sites in the seed region of the miRNAs were pointed out by “*“. b, c Luciferase reporter assays were performed using miR-15b (b) or miR-322 (c) co-transfected with a dual-luciferase reporter construct containing different lengths of 3′-UTR of SETD3. WT wild- type, x1 a single 3′-UTR sequence, x3 triple 3′-UTR repeat sequences. d The luciferase reporter constructs containing different 3′-UTR fragments of SETD3 were transfected into C2C12 cells stably expressing pri-miR-15b or pri-miR-322 construct. Luciferase activities from the indicated cells were measured. FL the full length of the 3′-UTR fragment (756 bp). e Relative expression levels of miR-15b and miR-322 shown in d were measured by RT-qPCR. f Luciferase reporter assays were performed using the indicated miRNA inhibitors co-transfected with a dual-luciferase reporter construct containing short length of 3′-UTR of SETD3. The relative luciferase activities were quantified from three biological repeats
MiR-15b and miR-322 repress SETD3 expression through binding to the 3′-UTR of SETD3
Next, we further explore whether miR-15b and miR-322 could repress SETD3 expression in vivo. Five different miRNA sense oligos were transfected into C2C12 cells and SETD3 transcriptional and protein levels were examined by real-time quantitative PCR (RT-qPCR) and Western blot analyses. Consistently, only miR-15b or miR-322 remarkably reduced SETD3 levels (Fig. 3a). Furthermore, we observed that transfection of the wild-type miRNA oligos, but not the miRNA-15b or miRNA-322 mutants in which either the seed sequences or the non-seed sequences have been changed, was able to affect SETD3 protein levels dramatically (Fig. 3b–d). The inert effect of miRNA-322 mutant 2 on SETD3 might result from its non-essential role of targeting 3′-UTR of SETD3 gene (Fig. 3d). As expected, overexpression of primary miR-15b or miR-322 construct in turn inhibited SETD3 protein levels (Fig. 3e). In addition, after antisense oligos of miR-15b or miR-322 (miRNA inhibitor) were transfected into C2C12 cells, both the transcriptional levels and the protein levels of SETD3 markedly increased compared to the control transfection (Fig. 3f, g). Alternatively, small guide RNA (sgRNA) was utilized to investigate the impact on SETD3 expression. The two sgRNAs were able to decrease expression level of miR-15b or miR-322, respectively (Fig. 3h). Consistently, SETD3 protein levels were increased by sgRNA knockdown of miRNAs, suggesting SETD3 may be the target of the two miRNAs in C2C12 cells (Fig. 3i). Consistent with previous reports that CCNE1 was also targeted by miR-15b and miR-322, the encoded Cyclin E1 protein levels were moderately increased with knockdown of these two miRNAs18,19,20,21. Importantly, the effect of two sgRNAs on SETD3 were not due to off-target effect, as coexpression of sgRNAs and their corresponding miRNA mimics compromised an accumulation of SETD3 caused by transfection sgRNAs alone (supplementary Fig. S2). Taken together, we conclude that miR-15b and miR-322 can directly target the 3′-UTR of SETD3 gene, which lead to inhibition of SETD3 expression.
a Transcriptional and protein levels of SETD3 in C2C12 cells expressing various miRNA mimics were examined by RT-qPCR (left panel) and western blot (right panel) analyses. b Sequences of the wild-type or mutants of miR-15b and miR-322 were shown. “*“ indicates the mutated sites. c, d SETD3 protein levels in cells expressing the indicated miRNA mimics or mutants shown in b were examined by western blot (left panel). The relative SETD3 levels normalized to β-Actin were quantified using ImageJ (right panel). e SETD3 protein levels in cells stably expressing the indicated pri-miRNAs were examined by western blot. f, g Transcriptional levels and protein levels of SETD3 in cells transfected the indicated miRNA inhibitors were examined by RT-qPCR (f) and western blot analysis (g, left panel). The relative SETD3 protein levels were quantified (g, right panel). h, i Transcriptional levels of miRNAs in cells transfected with sgRNA targeting miR-15b or miR-322 were examined by RT-qPCR in h, and protein levels of SETD3 were examined by western blot in i. The error bars in each quantification plot represent mean ± SD from three biological repeats
MiR-15b and miR-322 repress myoblast differentiation
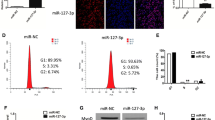
Since both miR-15b and miR-322 can repress SETD3 expression, we want to determine how these two miRNAs are expressed during muscle cell differentiation. To do this, we first utilized a database published previously and analyzed the expression levels of these miRNAs in various mouse tissues22. Consistently, the expression level of miR-206, a muscle-specific miRNA, is enhanced over 2000 fold in mouse muscle compared with that in mouse embryonic stem (ES) cells10. Interestingly, the expression levels of miR-15b and miR-322 are significantly decreased in mouse muscle compared to those in ES cells (33-fold or 8-fold reduction respectively) (Fig. 4a and supplementary Table S1). These two miRNAs are comparably expressed in other tissues, suggesting that they are not muscle-specific miRNAs. To this end, we then investigated the dynamic expression levels of miR-15b and miR-322 during C2C12 differentiation by RT-qPCR. MiR-1 was served as a positive control, as its expression level has been reported to gradually increase during muscle differentiation8; whereas miR-16 was served as a negative control, as it does not target SETD3 demonstrated in our result (Fig. 1b, c). We observed that miR-15b levels were declined during C2C12 differentiation, which is inversely correlated with SETD3 levels. Intriguingly, miR-322 levels remained unchanged, which is consistent with the results that we analyzed using the public GEO database (NCBI, Gene Expression Omnibus, www.ncbi.nih.gov/geo) (Fig. 4b). Next, undifferentiated C2C12 cells were transfected with synthetic miRNAs mimics, and treated with differentiation medium to induce myogenic differentiation for 4 days. RT-qPCR analysis showed that endogenous SETD3 mRNA levels were decreased (Fig. 4c). Western blot analysis confirmed that the protein levels of differentiation markers, such as MHC and Myogenin, decreased upon transfection of these two miRNAs mimics (Fig. 4d). Furthermore, immunofluorescence assays showed that myoblasts transfected with those miRNA mimics attenuated myoblast differentiation, as visualized by a significant decrease in the number and size of myotubes (Fig. 4e, f). Of note, the nuclei numbers per fiber were dramatically decreased, illustrating the defect of myoblast fusion into myotubes (Fig. 4g). Thus, we concluded that miR-15b and miR-322 may function in repression of myoblast differentiation.
a The expression levels of the indicated miRNAs in embryonic stem (ES) cells and various mouse tissues were plotted using the datasets in GEO. b The expression levels of the indicated miRNAs during C2C12 cells differentiation were analysed by RT-qPCR (top panel) or by using the datasets in GEO (bottom panel). c–g C2C12 cells were transfected with the indicated miRNA mimics, and induced differentiation for 4 days. c The mRNA levels of SETD3 and CCNE1 from differentiated cells were examined by RT-qPCR. d The differentiation protein markers in the transfected cells were analyzed by western blot analysis. e Immunofluorescence staining was performed using a specific antibody against MHC (red). DAPI stains nuclei. Scale bar: 100 μm. f MHC-positive cell numbers in certain field of the indicated C2C12 cells describe in e were quantified. g Quantitative analyses of nuclei number per fiber from the indicated C2C12 cells. Data represent the mean ± SD from three independent experiments
MiR-15b and miR-322 inhibit SETD3 to regulate muscle cell differentiation
Next we determine whether reduction of miR-15b or miR-322 could derepress its negative role in muscle differentiation. Therefore, a pair of sgRNAs or miRNA inhibitors that targeted miR-15b or miR-322, respectively, was transfected into C2C12 cells. After 4 days induction of myogenic differentiation, a significant increase of SETD3 levels accompanied with elevated levels of myogenic markers MHC and Myogenin were observed (Fig. 5a, b). Furthermore, immunofluorescence assays showed that myoblasts transfected with miRNA inhibitors formed more myotubes (Fig. 5c). Quantitative measurement of the numbers of myotubes and the nuclei numbers per fiber demonstrated that repression of miR-15b or miR-322 promotes myoblast differentiation (Fig. 5d, e).
a, b C2C12 cells transfected with the indicated sgRNAs (a) or miRNA inhibitors (b) targeting miR-15b or miR-322 were differentiated for 4 days, and the levels of the indicated proteins were examined by western blot. c Immunofluorescence analyses of the indicated differentiated C2C12 cells were stained with antibodies against SETD3 (green) and MHC (red). DAPI stains nuclei. Scale bar: 100 μm. d MHC-positive cell numbers in certain field of the indicated C2C12 cells describe in c were quantified. e Quantitative analyses of nuclei number per fiber from the indicated C2C12 cells. Data represent the mean ± SD from five fields. f Knockdown of SETD3 rescues miR-15b or miR-322 mediated myoblast differentiation. The protein levels of MHC and SETD3 were examined by western blot. S.E. short exposure, L.E. long exposure
To validate whether miR-15b or miR-322 affects myoblast differentiation via regulating SETD3, a rescue experiment was performed. C2C12 cells were transfected with siRNA targeting SETD3. After 4 h treatment, the inhibitor of miR-15b or miR-322 was separately added into cells for additional 6 h. After continued to culture in fresh media for 24 h, cells were induced to differentiation for additional 4 days. Inhibition of endogenous miR-15b or miR-322 increased SETD3 levels and facilitated cell differentiation; whereas knockdown of endogenous SETD3 rescued miR-15b or miR-322 inhibitors-mediated cell differentiation, evaluated by MHC levels (Fig. 5f). This result suggested that miR-15b and miR-322 repress muscle cell differentiation via inhibition of SETD3 expression.
E2F1 and FAM3B regulate SETD3 levels through controlling miR-15b or miR-322 expression, respectively
Next, we want to address how expressions of miR-15b and miR-322 are regulated during muscle differentiation. Previous studies have demonstrated that the pivotal transcription factor E2F1 directly targets promoters of miR-15 and miR-16 clusters and E2F1 inhibits myogenic differentiation23,24. Meanwhile, Zhang et al. recently reported that FAM3B inhibits miR-322 expression during high glucose induced vascular smooth muscle cell proliferation25. These results prompt us to investigate whether E2F1 and FAM3B regulate expression of miR-15b and miR-322 during skeletal muscle differentiation, respectively. To test this, we first examined the expression profiles of E2F1 and FAM3B. As expected, during myogenic differentiation process, E2F1 expression was reduced, whereas FAM3B expression was gradually increased (Fig. 6a). Consistently, knockdown of the positive transcriptional regulator E2F1 remarkably repressed miR-15b expression, and consequently increased SETD3 expression, but had no obvious impact on miR-322 expression (Fig. 6b). Meanwhile, knockdown of the negative regulator FAM3B promoted miR-322 expression, and consequently reduced SETD3 expression, but had no effect on miR-15b expression (Fig. 6c).
a The transcriptional levels of the indicated genes during muscle differentiation were examined by RT-qPCR. b, c The transcriptional levels of miR-15b or miR-322 after knockdown E2F1 (b) or knockdown FAM3B (c) were examined. The relative SETD3 levels were shown. d, e Western blot assays were performed to examine SETD3 levels after overexpression of E2F1 (d, left panel) or FAM3B (e, left panel) in C2C12 cells. Data represent the mean ± SD from three independent experiments (right panel). f Knockdown of FAM3B by two different siRNAs reduces SETD3 levels. g, i C2C12 cells were transfected with siRNAs targeting E2F1 or FAM3B, respectively, and induced to differentiate for 3 days. The indicated miRNAs or mRNA levels were examined by RT-qPCR. Data represent the mean ± SD from three independent experiments. h, j The same set of cells described in panel g, i were subjected to immunoblotting against the indicated antibodies. S.E. short exposure, L.E. long exposure
We speculate that E2F1 and FAM3B could regulate muscle differentiation by affecting SETD3 levels. Thus, SETD3 levels were examined in C2C12 cells transfected with HA-tagged E2F1 by immunoblotting. Interestingly, overexpression of E2F1 reduced SETD3 protein level (Fig. 6d). Similarly, when we altered FAM3B levels by overexpression or siRNA knockdown FAM3B, SETD3 protein levels were accordingly changed as expected (Fig. 6e, f). Moreover, if E2F1 was inhibited by siRNA knockdown, we observed much faster cell differentiation compared to the control cells after switching cells to the differentiation medium (Fig. 6g, h). In contrast, knockdown of FAM3B slowed down cell differentiation compared to the control cells, as both SETD3 and Myogenin levels were obviously decreased in same differentiation conditions (Fig. 6i, j). Together, these data indicated that E2F1 or FAM3B either positively or negatively regulates miRNAs, consequently affects SETD3 and muscle differentiation.
Discussion
In this study, we uncover a novel function of miR-15b and miR-322 in C2C12 differentiation beyond their roles in cancers26,27. Furthermore, we verify that a well-known transcription factor E2F1 is required for the reduction of miR-15b expression and the upregulation of SETD3, thereby promoting myoblast transition to myotube formation. Meanwhile, we also illustrate that a negative transcription regulator FAM3B is upregulated during this differentiation process, accompanied with decreased miR-322 level as well as increased SETD3 level (Fig. 7). These two parallel pathways of regulation of SETD3 expression highlight the importance of a protein-miRNAs interplay network during skeletal muscle differentiation.
MiRNAs have been elucidated to participate in almost every aspect of biology. For instance, the miR-15a/16-1 and miR-15b/16-2 clusters have been shown to regulate cell cycle and apoptosis by targeting CCND3 or CCNE124. MiR-15b has been shown to play roles in adipogenesis, lipid metabolism, and modulating DNA damage response28,29,30. In addition, miR-15a and miR-15b also functions as tumor suppressors, especially in B-cell oncogenesis, suggesting their potential clinical application26,31. It is worthy to note here that we uncovered miR-15b, but not miR-15a, regulates skeletal muscle differentiation, although these two miRNAs share very similar sequences. We have provided clear evidence showing that the unique role of miR-15b in muscle differentiation (Figs. 1c and 3c). Given that miR-15a and miR-15b are located at different chromosome loci, it is conceivable that these two miRNAs have distinct roles32.
Unlike miR-15b, miR-322 has been indicated to regulate muscle differentiation as well as cardiomyocyte specification33,34. As an X-chromosome miRNA, miR-322/-503 cluster specifically drives a cardiomyocyte program meanwhile inhibiting neural lineages33. MiR-322 can promote osteoblast differentiation by downregulation of Tob2 and Tob2-regulated osteogenic genes34. Meanwhile, miR-322 represses muscle differentiation, as overexpression of miR-322 mimics dampened myotube formation but promoted bone formation34. Consistently, we show here that inhibition of miR-322 significantly accelerate myotube formation, further confirming its negative role in myoblast differentiation (Fig. 5). In contrast, miR-322 can promote cell cycle quiescence and differentiation by down-regulation of Cdc25A27. Despite of this, whether miR-322 represses muscle differentiation has not been examined in that study. Our data have demonstrated that miR-322 indeed represses muscle differentiation. Nevertheless, why constant transcript levels of miR-322 are sustained during myoblast differentiation awaits further investigation.
It is interesting to understand how expression of the two miRNAs themselves is regulated during muscle differentiation. Here we provide evidence showing that E2F1 or FAM3B regulates expression of miR-15b or miR-322, respectively, during this process: (1) expression of E2F1 and FAM3B are dynamically altered from myoblast state to myotube formation; (2) E2F1 and FAM3B specifically control expression of the two miRNAs through directly targeting their corresponding promoters, respectively; (3) knockdown of FAM3B in C2C12 cells results in decreased SETD3 expression, which are correlated with repression of muscle cell differentiation (Fig. 6). Actually, previous studies have shown many clues to support our findings. First, E2F1-mediated transcription plays an essential role in muscle differentiation and myogenesis23,35. E2F1 expression is irreversibly downregulated during C2C12 myoblast differentiation, whereas overexpression of E2F1 promotes myoblast proliferation and represses myogenic differentiation23,36. Second, E2F family members, including E2F1 and E2F3, can directly bind to the promoter of miR-15b-16-2, and positively regulate miRNA expression during cell proliferation24,37. In addition, FAM3B protein is significantly increased during the proliferation and migration of vascular smooth muscle cells, accompanied with the inhibition of miR-322-5p, linking FAM3B to miR-322 regulation25. Moreover, luciferase reporter assay has been shown that FAM3B represses transcription of miR-322 by binding the promoter of miR-32225. Therefore, we at the first time demonstrate that E2F1 and FAM3B can regulate SETD3 through two parallel miRNA regulatory pathways, and decipher a complex network during myogenic differentiation.
Using cultured myoblast cell system, our current studies convincingly demonstrate the function and regulation of miR-15b and miR-322 in myoblast differentiation. It will be important to determine whether the repression of SETD3 by miRNAs contributes to skeletal muscle development and function. It will also be interesting to determine if miR-15b/miR-322 and SETD3 participate in skeletal muscle degeneration/regeneration process as well as human muscular diseases, such as rhabdomyosarcoma.
Methods and Materials
MiRNA prediction
MiRNAs potentially targeting the 3′-UTR of SETD3 gene were predicted by the TargetScan (http://www.targetscan.org/mmu_71/) and the Miranda (http://34.236.212.39/microrna/home.do) websites.
Construction of plasmids
The E2F1 and FAM3B from human cDNA library were transferred to pCS2-based Gateway vector containing 3xHA tag via LR reaction as described previously38.
Cell culture and transfection
C2C12 mouse myoblasts were cultured in growth medium (GM) — DMEM containing 20% fetal bovine serum (FBS) and maintained in a humidified incubator with 5% CO2 at 37 °C. For myogenic differentiation, when confluence was reached to 80–90%, C2C12 cells were shifted into a differentiation medium — DMEM containing 2% horse serum (HS). 293 T cells were cultured in DMEM containing 10% FBS and maintained in a humidified incubator with 5% CO2 at 37 °C. For miRNAs and plasmids transfection, when cells reached 60–70% confluence, the miRNAs or plasmids were transfected by the transfection reagent MAX according to the manufacturer’s protocol. Cells were harvested in 36–48 h after transfection of plasmids or 48–96 h after transfection of siRNAs or miRNAs. Unless stated, 293T cells were only used for the luciferase reporter assays; C2C12 cells were mainly used for cell differentiation experiments.
The synthesized miRNA or siRNA sequences (GenePharma Com. from Shanghai) are below: miR-15b mimics (WT): 5′- cagcagcacauaucagguuuaca-3′; miR-15b mutant 1: 5′-cucgucgacaucaugguuuaca-3′; miR-15b mutant 2: 5′-cagcagcacauguagguuuaca-3′; miR-322 mimics (WT): 5′-cagcagcaauucauguuuugga-3′; miR-322 mutant 1: 5′-cucgucguuuucauguuuugga-3′; miR-322 mutant 2: 5′-cagcagcaauuguaguuuugga-3′; miR-15b inhibitor: 5′-UGAA- CCAUGAUGUGCUGCUA-3′; miR-322 inhibitor: 5′-UCCAAAACAUGAAUUGCUGCUG-3′; si-mE2F1-1: 5′-ATCTGACCACCAAACGCTT-3′; si-mE2F1-2: 5′-GCCCTTGACTATCACTTTGGT-3′; si-mFAM3B-1: 5′-CAAACTGAAGGCTCAAGCAAA-3′; si-mFAM3B-2: 5′-GCACTCTCTACAACATCGAA-3′.
Western blot
Cells were lysed by RIPA buffer and added the bromophenol blue loading buffer, and then the samples were boiled for 10 min and centrifuged at 12,000 rpm for 5 min. The whole-cell lysate was separated into 8% SDS-acrylamide gels and transferred to PVDF membranes. After that, the membranes was blocks by 5% milk in TBST and probed with primary antibodies including mouse SETD3 (3B3, generated by Wuhan Dia-An Company), rabbit polyclonal SETD3 (Abclonal, A8071), MyoD1 (Proteintech, 18943-l-AP), HDAC1 (Abclonal, A2238), Cyclin E1 (Cell Signaling Technology, 20808 S), Myogenin (Abcam, ab124800; or Santa Cruz, D-10, sc-13137), MHC (Developmental Studies Hybridoma Bank, MF-20), β-Actin (Proteintech, 6008-I-Ig), and α-Tubulin (Sigma, T9026). For generation of mouse monoclonal SETD3 antibody, His-tagged full-length human SETD3 protein was expressed in E. Coli and purified as described previously14. Purified His-SETD3 proteins were immunizated into 5-8 weeks old Balb/C mice and boosted additional 4 times. After several steps including hybridoma production, screening, cloning, and expanding the hybridomas, a subclone named 3B3 was validated and amplified followed the procedure described as before39. Membranes were further probed with horseradish peroxidase (HRP)-conjugated secondary antibodies and the protein bands were visualized using chemiluminescence detection reagents.
RNA extraction, reverse transcription, and real-time quantitative PCR
Total RNA was isolated from C2C12 cells with TRIzol (Life technologies). The mRNA reverse transcription and real-time PCR were according to the manufacturer’s protocol (TIANGEN). The miRNA reverse transcription and real-time PCR were using the Hairpin-itTM Real-Time PCR Kit (Shanghai GenePharma). The primer sequences used in RT-qPCR are available upon request.
Dual-luciferase reporter assays
The dual-luciferase reporter plasmid psiCHECK2 was generously gifted from Xiang-Dong Fu laboratory. The longer 3′-UTR fragment of SETD3 gene was amplified by PCR from cDNA of C2C12 cells and cloned into psiCHECK2 vector’s downstream of the stop codon of Renilla luciferase gene. For Luciferase reporter assays, 20 nM miRNA and 10 ng plasmid were transfected into 293 T cells or C2C12 cells. After 24–48 h, cells were lysed and the luciferase activity was tested according to the manufacturer’s instructions (Promega).
Generation of SETD3 knockdown cell line
Short hairpin RNA fragments (shRNAs) of SETD3 containing 5′- CATCACCATGTTCCTTGTTAA-3′ (shSETD3-1) or 5′-GCTGGAGATCA- GATTTACATT-3′ (shSETD3-2) were cloned into plko.1 vector using the restriction enzymes EcoRI and AgeI (New England Biolabs). To obtain lentivirus, the knockdown plasmids were transfected into 293 T cells along with the helper plasmids pMD2G and psPAX2 using the ratio of 2:1:1. Cell culture medium was changed after 12 h transfection and virus were harvested 24 h later with filter. Cells were seeded into a 12-well plate 1 day before lentivirus infection. SETD3 knockdown cells will be harvested after 36–48 h.
Knockdown of miRNAs by CRISPR-Cas9 technology
We designed two sgRNAs each miRNA by the CRISPR Design Tool (http://tools.genome-engineering.org) and inserted them into pSpCas9 (BB)-2A-Puro vector. The sgRNA sequences are below: miR-15b-sgRNA-1: 5′-AGTACTGTAGCAGCACATCA-3′; miR-15b-sgRNA-2: 5′-CAAACATAATACAACTGTGA-3′; miR-322-sgRNA-1: 5′-CCCTTCGGAGTCAACGAGGG-3′; miR-322-sgRNA-2: 5′-GCGCTGCAACACCCCTTCGT-3′. After CRISPR-Cas9 plasmids transfected and selected by puromycin for 2-3 days, C2C12 cells were harvested and SETD3 expression level was analyzed by western blot.
Pri-miRNA overexpression system
Pri-miRNA sequences were searched from UCSC Genome Browser (http://genome.ucsc.edu) and a nucleotide segment containing mi-15b or mi-322 was cloned into pHAGE-CMV vector using the restriction enzymes NotI and XhoI (New England Biolabs). The primers used for construction of pri-miRNA are as follows: pri-miR-15b forward (F): 5′-ATAAGAATGCGGCCGCGCCACCGGCATTG-ACTTAGACCATAATC-3′; pri-miR-15b reverse (R): 5′-CCGCTCGAGCACTACGCCAATATTTACGTG- 3′; pri-miR-322 forward (F): 5′-ATAAGAATGCGGCCGCGCCACCCTGAGGTAAGAGTCTCCTCC-3′; pri-miR-322 reverse (R): 5′-CCGCTCGAGGTGACCCTCACTAGACTAA-G-3′. 293 T cells were infected and selected according to the lentiviral expression and packaging protocol described above. The packaged virus was used to infect C2C12 cells to generate pri-miRNA stably expressed cell lines.
Immunofluorescence staining
C2C12 cells were cultured on glass coverslips, induced to differentiation for 4 days, fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 (Sigma) for 10 min, blocked with 3% BSA solution, incubated with an primary antibody (for MHC: 1:50; for SETD3 1:100) at 4 °C overnight, incubated with a secondary antibody at room temperature for 1 h. The coverslips were stained with DAPI and mounted. Immunofluorescence images were captured under a confocal laser-scanning microscope (Leica SP8).
Statistical analysis
For quantification of the western blot data, ImageJ software was used to measure the relative intensity of each band. Data are presented as mean ± standard deviation (SD) from at least three biological replicates, and the difference between any two groups were compared by Student’s t-test using Prism 5 software. NS not significance, *p < 0.05, **p < 0.01, ***p < 0.001.
References
Rudnicki, M. A., Le Grand, F., McKinnell, I. & Kuang, S. The molecular regulation of muscle stem cell function. Cold Spring Harb. Symp. Quant. Biol. 73, 323–331 (2008).
Buckingham, M. & Rigby, P. W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 28, 225–238 (2014).
Fong, A. P. & Tapscott, S. J. Skeletal muscle programming and re-programming. Curr. Opin. Genet. Dev. 23, 568–573 (2013).
Berkes, C. A. & Tapscott, S. J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16, 585–595 (2005).
McKinsey, T. A., Zhang, C. L. & Olson, E. N. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14, 763–772 (2002).
Taylor, M. V. & Hughes, S. M. Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 72, 33–44 (2017).
Luo, W., Nie, Q. & Zhang, X. MicroRNAs involved in skeletal muscle differentiation. J. Genet. Genom. 40, 107–116 (2013).
Chen, J. F. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 (2006).
McDaneld, T. G. et al. MicroRNA transcriptome profiles during swine skeletal muscle development. BMC Genom. 10, 77 (2009).
Kim, H. K., Lee, Y. S., Sivaprasad, U., Malhotra, A. & Dutta, A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 174, 677–687 (2006).
Eom, G. H. et al. Histone methyltransferase SETD3 regulates muscle differentiation. J. Biol. Chem. 286, 34733–34742 (2011).
Wilkinson, A. W. et al. SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature 565, 372–376 (2019).
Kwiatkowski S., et al. SETD3 protein is the actin-specific histidine N-methyltransferase. Elife 7, e37921 (2018).
Cheng, X. et al. Cell cycle-dependent degradation of the methyltransferase SETD3 attenuates cell proliferation and liver tumorigenesis. J. Biol. Chem. 292, 9022–9033 (2017).
Balkhi, M. Y. et al. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci. Signal. 6, ra63 (2013).
Antoniou, A., Mastroyiannopoulos, N. P., Uney, J. B. & Phylactou, L. A. miR-186 inhibits muscle cell differentiation through myogenin regulation. J. Biol. Chem. 289, 3923–3935 (2014).
Han, C. et al. miR-29a promotes myocardial cell apoptosis induced by high glucose through down-regulating IGF-1. Int. J. Clin. Exp. Med. 8, 14352–14362 (2015).
Xia, H. et al. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem. Biophys. Res. Commun. 380, 205–210 (2009).
Vimalraj, S. & Selvamurugan, N. Regulation of proliferation and apoptosis in human osteoblastic cells by microRNA-15b. Int. J. Biol. Macromol. 79, 490–497 (2015).
Ichihara, A. et al. microRNA-mediated keratinocyte hyperproliferation in psoriasis vulgaris. Br. J. Dermatol. 165, 1003–1010 (2011).
Nakashima, T. et al. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: its implications to therapy. PLoS ONE 5, e14334 (2010).
Kuchen, S. et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32, 828–839 (2010).
Wang, J., Helin, K., Jin, P. & Nadal-Ginard, B. Inhibition of in vitro myogenic differentiation by cellular transcription factor E2F1. Cell Growth Differ. 6, 1299–1306 (1995).
Ofir, M., Hacohen, D. & Ginsberg, D. MiR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol. Cancer Res. 9, 440–447 (2011).
Zhang, W., Chen, S., Zhang, Z., Wang, C. & Liu, C. FAM3B mediates high glucose-induced vascular smooth muscle cell proliferation and migration via inhibition of miR-322-5p. Sci. Rep. 7, 2298 (2017).
Aqeilan, R. I., Calin, G. A. & Croce, C. M. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 17, 215–220 (2010).
Sarkar, S., Dey, B. K. & Dutta, A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol. Biol. Cell 21, 2138–2149 (2010).
Dong, P. et al. MiR-15a/b promote adipogenesis in porcine pre-adipocyte via repressing FoxO1. Acta Biochim. Biophys. Sin. 46, 565–571 (2014).
Chu, M. et al. miR-15b negatively correlates with lipid metabolism in mammary epithelial cells. Am. J. Physiol. Cell Physiol. 314, C43–C52 (2018).
Rahman, M. et al. miR-15b/16-2 regulates factors that promote p53 phosphorylation and augments the DNA damage response following radiation in the lung. J. Biol. Chem. 289, 26406–26416 (2014).
Lovat, F. et al. miR-15b/16-2 deletion promotes B-cell malignancies. Proc. Natl Acad. Sci. USA 112, 11636–11641 (2015).
Calin, G. A. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA 99, 15524–15529 (2002).
Shen, X. et al. miR-322/-503 cluster is expressed in the earliest cardiac progenitor cells and drives cardiomyocyte specification. Proc. Natl Acad. Sci. USA 113, 9551–9556 (2016).
Gamez, B., Rodriguez-Carballo, E., Bartrons, R., Rosa, J. L. & Ventura, F. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. J. Biol. Chem. 288, 14264–14275 (2013).
Wang, J., Huang, Q., Tang, W. & Nadal-Ginard, B. E2F1 inhibition of transcription activation by myogenic basic helix-loop-helix regulators. J. Cell Biochem. 62, 405–410 (1996).
Luo, W., Li, G., Yi, Z., Nie, Q. & Zhang, X. E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation. Sci. Rep. 6, 27904 (2016).
Bueno, M. J. et al. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Mol. Cell Biol. 30, 2983–2995 (2010).
Wang, H. Y., Li, Y., Xue, T., Cheng, N. & Du, H. N. Construction of a series of pCS2 + backbone-based Gateway vectors for overexpressing various tagged proteins in vertebrates. Acta Biochim. Biophys. Sin. 48, 1128–1134 (2016).
Foung, S. K., Engleman, E. G. & Grumet, F. C. Generation of human monoclonal antibodies by fusion of EBV-activated B cells to a human-mouse hybridoma. Methods Enzymol. 121, 168–174 (1986).
Acknowledgements
We are grateful to Drs. Xi Zhou (Wuhan Institute of Virology, Chinese Academy of Sciences) and Yu Zhou (Wuhan University) for plasmids and technical help. We also thank Dr. Zhenji Gan (Nanjing University) for discussion and Ms. Yumin Li and Hongguo Duan for technical assistant. This work was supported by the Major State Basic Research Development Program of China (2013CB910700 to H.N.D.), the National Natural Science Foundation of China (31770843 and 31271369 to H.N.D.), and Wuhan University (2042018kf0217 to W.J. and H.N.D.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Calin
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, MJ., Xie, J., Shu, WJ. et al. MiR-15b and miR-322 inhibit SETD3 expression to repress muscle cell differentiation. Cell Death Dis 10, 183 (2019). https://doi.org/10.1038/s41419-019-1432-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-1432-5
- Springer Nature Limited
This article is cited by
-
miR-322 promotes the differentiation of embryonic stem cells into cardiomyocytes
Functional & Integrative Genomics (2023)
-
Stabilization of SETD3 by deubiquitinase USP27 enhances cell proliferation and hepatocellular carcinoma progression
Cellular and Molecular Life Sciences (2022)
-
Evaluation of muscle-specific and metabolism regulating microRNAs in a chronic swimming rat model
Journal of Muscle Research and Cell Motility (2022)
-
miR-322/-503 rescues myoblast defects in myotonic dystrophy type 1 cell model by targeting CUG repeats
Cell Death & Disease (2020)
-
Fetal malnutrition-induced catch up failure is caused by elevated levels of miR-322 in rats
Scientific Reports (2020)