Abstract
Dysregulation of long noncoding RNAs (lncRNAs) has been associated with the development and progression of many human cancers. Lactate dehydrogenase A (LDHA) enzymatic activity is also crucial for cancer development, including the development of papillary thyroid cancer (PTC). However, whether specific lncRNAs can regulate LDHA activity during cancer progression remains unclear. Through screening, we identified an LDHA-interacting lncRNA, GLTC, which is required for the increased aerobic glycolysis and cell viability in PTC. GLTC was significantly upregulated in PTC tissues compared with nontumour thyroid tissues. High expression of GLTC was correlated with more extensive distant metastasis, a larger tumour size, and poorer prognosis. Mass spectrometry revealed that GLTC, as a binding partner of LDHA, promotes the succinylation of LDHA at lysine 155 (K155) via competitive inhibition of the interaction between SIRT5 and LDHA, thereby promoting LDHA enzymatic activity. Overexpression of the succinylation mimetic LDHAK155E mutant restored glycolytic metabolism and cell viability in cells in which metabolic reprogramming and cell viability were ceased due to GLTC depletion. Interestingly, GLTC inhibition abrogated the effects of K155-succinylated LDHA on radioiodine (RAI) resistance in vitro and in vivo. Taken together, our results indicate that GLTC plays an oncogenic role and is an attractive target for RAI sensitisation in PTC treatment.
Similar content being viewed by others
Introduction
Papillary thyroid cancer (PTC) is the most common histological type of thyroid cancer (accounting for 89.1% of cases) [1]. The incidence of thyroid cancer has increased significantly over the past few decades, driven primarily by increases in the diagnosis of this PTC subtype [2]. PTC, a well-differentiated subtype, has been reported to be able to progress to a poorly differentiated status and become refractory to radioiodine (RAI), with a rapid decrease in life expectancy [3, 4]. Since the molecular mechanisms of PTC progression are not fully understood, more research is needed to discover novel risk factors and therapeutic targets.
Hyperglycolysis is a hallmark of cancer [5]. One predominant feature of poorly differentiated PTC is active glucose metabolism, which can be visualised by the high uptake of 18F-fluorodeoxyglucose (18F-FDG), a glucose analogue radiotracer, on positron emission tomography/computed tomography (PET/CT) imaging [6]. Patients with lesions exhibiting positive 18F-FDG uptake might derive little benefit from RAI treatment [7]. Increased aerobic glycolysis often correlates with tumour aggressiveness and poor prognosis. However, the molecular mechanisms underlying glycolysis in PTC remain elusive.
Lactate dehydrogenase A (LDHA) is a crucial enzyme in aerobic glycolysis. LDHA activity is essential for maintaining a high glycolytic rate by regenerating NAD+, which is required in the early steps of glycolysis [8]. LDHA produces lactate to acidify the extracellular pH in the tumour microenvironment and promotes many processes, such as metastasis, angiogenesis and immunosuppression, which are associated with a worse clinical prognosis [9]. Furthermore, lactate can be taken up by self or adjacent tumour cells under nutrient-limited conditions and then used as a substrate for a downstream pathway (mitochondrial respiration) [10]. Thus, LDHA and its cometabolite play a central role in tumour metabolic reprogramming.
Long noncoding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides with little or no protein-coding capacity [11]. Recent studies have separately investigated the roles of lncRNAs and metabolic reprogramming mediated by several glycolytic enzymes and regulators in promoting tumorigenesis and tumour progression [12,13,14,15,16,17]. The crosstalk between lncRNAs and the establishment of metabolic reprogramming in PTC cells and the related potential mechanisms remain poorly understood and thus attracted our interest.
Materials and methods
Cells and reagents
BCPAP, KTC-1 and HEK293T cells were purchased from the National Collection of Authenticated Cell Cultures. TPC-1, K1, and Nthy-ori 3-1 cells were purchased from Shanghai Qida Biotechnology Co., Ltd. Although BCPAP, TPC-1, KTC-1, and K1 cells are human papillary thyroid carcinoma-derived cells, KTC-1 cells are functionally poorly differentiated [18, 19]. Nthy-ori 3-1 is a normal human thyroid cell line. BCPAP, KTC-1, TPC-1, and Nthy-ori 3-1 cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% foetal bovine serum (Gibco). K1 cells were cultured in Leibovitz’s L-15 medium (Gibco).
All cells were tested for and found to be free of mycoplasma contamination. The antibodies used were as follows: anti-sirtuin 5 (SIRT5; catalogue #15122-1-AP, Proteintech, Rosemont, IL, USA), anti-HA (catalogue #H9658, Sigma‒Aldrich, St. Louis, MO, USA), anti-Flag M2 (catalogue #F1804, Sigma‒Aldrich), anti-Ki67 (catalogue #ab16667, Abcam, Cambridge, MA, USA), anti-β-actin (catalogue #4970, Cell Signaling Technology, Boston, MA, USA), anti-acetylated lysine (catalogue #9681, Cell Signaling Technology), anti-succinyllysine (catalogue #PTM-401, PTM Biolabs, Hangzhou, China), anti-glutaryllysine (catalogue #PTM-1151, PTM Biolabs), and anti-malonyllysine (catalogue #PTM-901, PTM Biolabs). Antibodies specifically recognising succinylation at lysine 155 (K155) of LDHA were prepared commercially at ChinaPeptides Co., Ltd. (Shanghai, China). The synthesised peptide ISGFP(Suc) NRVIGSGCN was coupled to keyhole limpet haemocyanin as an antigen for immunisation of rabbits. Antiserum was collected after four doses of antigen.
The GLTC, GLTC-MUT, Flag-SIRT5, HA-LDHAWT, HA-LDHAK155E, HA-LDHAK155R and empty vector sequences were subcloned into the expression vector pcDNA3.1 (Invitrogen, Waltham, MA, USA). All plasmids and siRNAs were transfected into PTC cells using Lipofectamine 2000 transfection reagent (Invitrogen, USA). The negative control shRNA (sh-NC) or GLTC shRNA (sh-GLTC) lentiviral vectors were transduced into cancer cells, as were the empty vector and GLTC overexpression lentiviral vectors. All plasmids, siRNAs and lentiviruses were purchased from GenePharma Technology (Shanghai, China). The sequences of the siRNAs are listed in Supplementary Table 1.
Clinical samples
Three independent cohorts of PTC patients were enrolled in this study. In cohort 1, PTC tissues and nontumor thyroid tissues (taken from a region 5 cm from the tumour edge) were obtained from four patients with advanced, aggressive PTC in November 2016 at the Affiliated Hospital of Jiangsu University, School of Medicine, Jiangsu University (Zhenjiang, China). In cohort 2, PTC tumour tissues and nontumor thyroid tissues were obtained from 44 patients who underwent surgery at the Affiliated Hospital of Jiangsu University between December 2016 and February 2017. In cohort 3, specimens from 82 PTC patients who underwent surgery were acquired from two hospitals: 72 from patients treated at the Affiliated Hospital of Jiangsu University between January 2007 and May 2016 and 10 from patients treated at Nanjing First Hospital, Nanjing Medical University (Nanjing, China) between August 2019 and August 2021. No patient had received any treatment prior to surgery. All clinicopathological information is provided in Supplementary Data 1, 2, and 3. The study protocol was approved by the ethics committees of the Affiliated Hospital of Jiangsu University and Nanjing First Hospital. Written informed consent was obtained from all participants in this study. All research was carried out in accordance with the provisions of the 1975 Declaration of Helsinki.
RNA extraction and quantitative RT–PCR (qRT‒PCR)
Total RNA was isolated with TRIzol reagent (Omega, Norcross, GA, USA) and transcribed to complementary DNA (cDNA) using a cDNA synthesis kit (Vazyme, Nanjing, China) according to the manufacturer’s protocol. qRT‒PCR was performed with gene-specific primers by using SYBR Green PCR Master Mix (TaKaRa, Shiga, Japan). The transcript levels of the genes were measured with a StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). The gene-specific primers are listed in Supplementary Table 1.
RNA sequencing (RNA-seq) analysis
Total RNA was extracted from four free-frozen PTC tissues and corresponding normal thyroid specimens (cohort 1) using TRIzol reagent (Invitrogen, USA). RNA quality was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). RNA-seq analysis was performed on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) with 150 bp paired-end reads. Illumina sequencing libraries were constructed using a TruSeq Stranded Total RNA Library Prep Kit (Illumina, USA) according to the manufacturer’s instructions. The differentially expressed genes were identified in samples of four paired PTC tissues and corresponding adjacent normal tissues. The expected fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) values were calculated to estimate lncRNA expression levels. LncRNA expression analysis was performed using the DESeq2 R package, which provides statistical routines to determine the differential expression of lncRNA molecules. LncRNAs with a corrected P value of <0.05 [false discovery rate <0.05] as calculated by the DESeq2 R package, a gene expression fold change of >2, and an average expression value of >5 were considered differentially expressed.
RNA immunoprecipitation (RIP) sequencing (RIP-seq)
RIP was performed using a Magna RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) according to the instructions. Cells (1 × 107 cells/100 mm dish) were lysed with one thousand microlitres of RIP lysis buffer containing a protease inhibitor cocktail and an RNase inhibitor. Washed magnetic beads were incubated with 5 μg of a human anti-LDHA antibody (catalogue # 19987-1-AP, Proteintech) or negative control IgG at room temperature for 30 min with rotation. Then, the cell lysate prepared as described above was added to the bead-antibody complexes. The mixture was incubated with rotation at 4 °C overnight. The samples were incubated with proteinase K to digest proteins, and immunoprecipitated RNA was isolated. Then, purified RNAs were reverse transcribed into cDNA. The cDNA libraries were sequenced on the Illumina sequencing platform and were subjected to RT–PCR to demonstrate the presence of the binding targets.
Subcellular fractionation assay
A PARIS™ Kit (Ambion, Austin, TX) was used to separate cytoplasmic and nuclear RNA. RNA isolated from the cytoplasmic and nuclear fractions was further analysed by qRT‒PCR. ACTB and GAPDH were used as cytoplasmic markers, and U6 was used as a nuclear marker.
Western blot analysis
Cells were lysed in RIPA buffer containing protease inhibitors by incubation for 30 min on ice, and the lysates were then centrifuged at 15,000 × g for 30 min at 4 °C. The samples were boiled and then analysed by western blotting as previously described [15].
LDHA activity assay
HA-LDHAWT, HA-LDHAK155E, and HA-LDHAK155R proteins were immunopurified from transfected cells, and LDHA activity was measured using an LDH Activity Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions.
In vitro 18F-FDG uptake assay, lactate production assay, and NAD+/NADH ratio determination
Cells were collected and washed three times with cold phosphate-buffered saline (PBS). Then, the collected cells were incubated in 1 mL of glucose-free culture medium containing 18F-FDG (111 kBq [3 μCi/mL]) for 1 h at 37 °C. The cells were then washed three times with cold PBS. One millilitre of 0.1 M NaOH was added to each well to obtain lysates. The radioactivity of the lysates was determined with a well γ-counter and normalised to the number of cells. For the lactate production assay, cells were washed with PBS and cultured in glucose-free culture medium for 12 h. The cells or tissues were lysed and centrifuged at 4 °C for 30 min. Then, the clear supernatants were collected for measurement of the lactate level according to the instructions of the Lactate Assay Kit (CMA, Sweden). A NAD+/NADH Quantitation Kit (Njjcbio, China) was used to quantify the NAD+/NADH ratio in the cell lysates. The values were normalised to the number of cells.
Cell viability and colony formation assays
Cell viability was assessed with a Cell Counting Kit-8 (Dojindo, Japan). A total of 5 × 103 cells per well were seeded in a 96-well plate. CCK-8 reagent (10 μl/well) was added to each well. After incubation for 1 h, the optical density at 450 nm (OD450) was measured for each sample.
For the plate colony formation assay, 300 cells per well were seeded in a 6-well plate. The medium was changed regularly. After culture for 14 days, cell colonies were stained with 0.1% crystal violet (1 mg/mL) and counted.
XF24 extracellular flux analysis
An XF24 extracellular flux analyser was used to evaluate the extracellular acidification rate (ECAR). In total, 2 × 104 cells per well were plated in XF24 cell culture plates (Seahorse Bioscience, USA) and incubated at 37 °C overnight. The ECAR was measured using a Seahorse XF Glycolysis Stress Test Kit (Agilent Technologies, USA). The values are presented as the means ± standard errors of the mean.
[6-14C]-Glucose CO2 release assay
Cells were cultured in glucose-free culture medium supplemented with 0.1 μCi/mL [6-14C]-glucose and incubated in a sealed chamber. The sealed chamber was placed in a 37 °C CO2-free cell incubator for 1 h. Then, the cells were treated with 500 μL of 1 M H2SO4. A centrifuge containing 1 mL of CO2 adsorbent phenylethylamine was placed in the sealed chamber. The sealed chamber was then placed in a CO2-free cell incubator at 37 °C for 1 h. The radiolabelled CO2 released from the cells was captured by phenylethylamine and measured with a scintillation counter.
Immunoprecipitation
For analysis of endogenous protein–protein interactions, five hundred microlitres of whole-cell lysates were incubated with an anti-SIRT5 antibody (catalogue #15122-1-AP, Proteintech, 1:50) or an anti-LDHA antibody (catalogue #19987-1-AP, Proteintech, 1:50) and 20 μL of protein A/G agarose (catalogue #QY2003, Pierce, Waltham, MA, USA) overnight at 4 °C. For the exogenous coimmunoprecipitation (co-IP) assay, five hundred microlitres of whole-cell lysates containing HA-tagged or Flag-tagged proteins were incubated with 20 µL of anti-HA agarose (catalogue #A2095 Sigma‒Aldrich) or anti-Flag (catalogue #A2220, Sigma‒Aldrich) affinity gel overnight at 4 °C. Then, sample loading buffer was added to the agarose and boiled. The resulting samples were analysed by western blotting.
In situ hybridisation (ISH) and immunohistochemical (IHC) analysis
ISH analysis of GLTC was performed in paraffin-embedded samples using a DIG-labelled miRCURYTM detection probe (Exiqon, Vedbaek, Denmark). The sequence of the GLTC probe was as follows: 5′–3′/5DigN/AGTTGAGCATCTCGTCGCCATTAC/3Dig_N/. In brief, sections were hybridised with a GLTC antisense probe overnight at 40 °C. Then, the sections were incubated with anti-DIG-AP Fab fragments for 60 min at 37 °C prior to conjugation with alkaline phosphatase for 30 min at 37 °C. Nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate colour substrate (NBT-BCIP; Roche, Germany) was used to detect the signals of the hybridised probes. A purple stain in the samples indicated a positive NBT-BCIP signal. The staining score for the ISH analysis was calculated by multiplying the percentage of positively stained tumour cells (0–100%) by the intensity of the staining (low, 1+; moderate, 2+; strong, 3+) [20]. Thus, the staining score ranged from 0 to 3. The median staining score was identified as the cut-off value. If the calculated score was below the median, the expression of the indicator was considered low; if the score was above the median, the expression of the indicator was considered high.
IHC analysis was performed as described previously [15]. The immunostaining intensity was classified into three grades: 0 = negative; 1 = weak; 2 = moderate; and 3 = strong. The distribution of staining was scored as follows: 1 ≤ 25% positive cells, 2 = 25–50% positive cells and 3 ≥ 50% positive cells. Thus, the final scores in those PTC tissues ranged from 0 to 9. A score of ≤3 was defined as low expression, and a score of >3 was defined as high expression.
RNA pulldown assay
LncRNAs were reverse transcribed with a MEGAscript T7 Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA) and biotinylated with a Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo Fisher Scientific) according to the manufacturers’ instructions. TPC-1 cells were collected and lysed in Pierce IP Lysis Buffer (Thermo Fisher Scientific). RNA pulldown assays were performed using a Pierce Magnetic RNA–Protein Pull-Down Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The full-length sense GLTC (F1), full-length antisense GLTC (F2), and GLTC truncation (Δ1: 1–250, Δ2: 1–500, Δ3: 251–627) probes were transcribed and biotinylated in vitro. TPC-1 cell lysates were incubated with these biotinylated RNA probes. Streptavidin magnetic beads were used to capture biotinylated RNA, and the biotinylated nucleic acid compounds were then incubated with cell lysates or purified proteins (20 µg) at 4 °C for 3 h. The proteins were eluted from the RNA-protein complexes and subjected to mass spectrometry (MS) analysis or western blotting.
Generation of LDHA knockout cell lines by CRISPR–Cas9 gene editing
To generate the LDHA-knockout (LDHA-KO) K1 cell lines, the sgRNA sequences (Supplementary Table 1) were ligated into the LentiCRISPRv2 plasmid. HEK293T cells were transiently transfected with the LentiCRISPRv2 plasmid and viral packaging and envelope plasmids (psPAX2 and pMD2.G, respectively). Forty-eight hours later, the viral supernatant was harvested and filtered through a 0.45 μm strainer. K1 cells were infected with the lentiviruses. Selection was carried out with puromycin (1 μg/mL) for 1 week. Infected cells were seeded as single cells (monoclonal culture) to obtain knockout cell lines, which were then verified by western blotting.
Xenograft mouse model and 18F-FDG micro-PET/CT
TPC-1 cells (1.0 × 107) stably expressing sh-NC or sh-GLTC were injected subcutaneously into 4-week-old female BALB/c nude mice. The mice were subjected to 18F-FDG PET scanning 21 days after injection. Specifically, mice were administered 18F-FDG (250 μCi per mouse) by tail vein injection after being fasted for 6 h. Forty-five minutes after 18F-FDG administration, the animals were anaesthetised with 2.5% isoflurane and immobilised during the 10 min PET scan acquisition (Inveon, Siemens). The regions of interest, which covered the entire tumour, were drawn around the tumours on CT scan slices. The maximal standard uptake values were calculated to assess the 18F-FDG uptake ability of the tumours. After PET/CT scanning, the mice were sacrificed, and the tumours were excised and weighed.
LDHA-KO K1 cells (1.0 × 107) stably expressing vector control or GLTC overexpression plasmid and the HA-LDHAWT, HA-LDHAK155R, or HA-LDHAK155E plasmid were injected subcutaneously into 4-week-old female BALB/c nude mice. All mice were killed 21 days after injection, and tumour tissues were collected and weighed.
131I-sensitive and 131I-resistant TPC-1 cells (1 × 107) stably expressing sh-NC or sh-GLTC were subcutaneously inoculated into 4-week-old female BALB/c nude mice. Fourteen days after subcutaneous inoculation, the mice were randomly divided into groups and intratumorally injected with 131I (2.8 mCi in 100 μL) or with saline as a control treatment. After 3 weeks of treatment, the mice were sacrificed, and the tumours were weighed. Sample sizes for all animal studies were selected based on expected effect size. Sample sizes (n) indicated in figure legends refer to the number of animal. The investigator was blinded to the group allocation and assessing the outcome during the experiments.
Fluorescence in situ hybridisation (FISH)
FISH analysis was performed using oligonucleotide-modified probes for human GLTC and a negative control probe with a lncRNA FISH Kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. In brief, PTC cells were grown on glass coverslips in 24-well plates. The cells were harvested, washed with PBS and fixed using 4% paraformaldehyde for 10 min. After permeabilization with 0.25% Triton X-100 for 10 min, the cells were washed with PBS and hybridised with FISH probes (designed by RiboBio) overnight in a humidified chamber at 37 °C in the dark. All images were acquired with a confocal laser scanning microscope (Zeiss, Oberkochen, Germany).
Liquid chromatography–tandem mass spectrometry (LC‒MS/MS) analysis of peptide succinylation
GLTC-transfected or control K1 cells were lysed in urea (8 M urea; 100 mM Tris/HCl, pH 8.5) buffer, and proteins were extracted. A Bradford Protein Assay Kit was used to quantify the amount of protein. A total of 20 µg of protein from each sample was mixed with 5× loading buffer and boiled for 5 min. The samples were subjected to 12.5% SDS‒PAGE followed by Coomassie Blue R-250 staining. Then, dithiothreitol (DTT; final concentration, 10 mM) was added to the samples, which were mixed at 600 rpm for 1.5 h (37 °C) and then cooled to room temperature. Iodoacetic acid (IAA; final concentration, 50 mM) was added to the mixture, which was incubated in the dark for 30 min. Four volumes of 50 mM Tris HCl (pH 8.0) was used to dilute the concentrate to 2 M. The samples were digested with trypsin (trypsin:protein (wt/wt) ratio, 1:50) at 37 °C overnight. The peptides were eluted from the beads with 0.1% trifluoroacetic acid (TFA) and were then combined and vacuum dried. The obtained peptides were cleaned with C18 cartridges (Empore™ SPE Cartridges C18 (standard density), 7 mm bed inner diameter [I.D.], 3 mL volume, Sigma) and lyophilised for further use.
The samples were dissolved in 1.4 mL of precooled IAP buffer and were then incubated with pretreated anti-succinyl-lysine antibody beads (PTMScan® Succinyl-Lysine Motif Kit, Cell Signaling Technology) for 1.5 h at 4 °C with gentle shaking, and the mixture was centrifuged at 2000 × g for 30 s. Then, the supernatant was discarded. The anti-succinyl-lysine antibody beads were washed 3 times with 1 mL of precooled IAP buffer and then 3 times with precooled water. The peptides were eluted from the beads with 40 μL of 0.1% TFA and were then combined and vacuum dried at room temperature. The peptides were added to 0.1% TFA again, and the mixture was centrifuged at 2000 × g for 30 s. The supernatant containing immune-enriched succinylated peptides was desalted on C18 stage tips.
LC‒MS/MS analysis was performed on a Q Exactive HF/HFX mass spectrometer (Thermo Fisher Scientific) coupled to an Easy nLC system (Proxeon Biosystems, now Thermo Fisher Scientific) for 120 min. The samples were loaded onto a reversed-phase trap column (Thermo Fisher Scientific, Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18) connected to a C18 reversed-phase analytical column (Thermo Fisher Scientific, Easy Column, 10 cm length, 75 μm I.D., 3 μm resin) in buffer A (0.1% formic acid) and eluted with a linear gradient of buffer B increasing from 5% solvent B (90% acetonitrile/0.1% formic acid, v/v) to 80% solvent B for 40 min at a flow rate of 300 nL/min. The mass spectrometer was operated in positive ion mode. MS data were acquired using a data-dependent top 10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for higher-energy C-trap dissociation (HCD) fragmentation. The automatic gain control target was set to 3e6, and the maximum injection time was set to 10 ms. The dynamic exclusion duration was set to 5 s. Survey scans were acquired at a mass resolution of 70,000 at m/z = 200, the resolution for HCD spectra was set to 17,500 at m/z = 200, and the isolation width was 2 m/z. The normalised collision energy was 30 eV, and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at the maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled. All MS/MS data obtained by LC‒MS/MS were combined and searched using MaxQuant software (version 1.3.0.5) for identification and quantitative analysis.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA) or SPSS 18.0 software (SPSS, Chicago, Ill., USA). The data are shown as the means ± SEMs, and unless otherwise indicated, all results are representative of at least three independent replicates. Two-tailed unpaired Student’s t test was used to analyse differences between two groups. One-way analysis of variance (ANOVA) was used to analyse differences among three or more groups. For clinicopathologic analysis, sample sizes were selected based on expected effect size. To analyse normality and homogeneity of data distribution, the Kolmogorov-Smirnov and Levene tests, respectively, were used. The chi-square test or Fisher’s exact test (two-sided) was performed. The survival curves were generated using the Kaplan‒Meier method, and differences were assessed by the log-rank test. Univariate and multivariate Cox regression analyses were used to determine the factors that influenced survival. P < 0.05 was considered statistically significant.
Results
The LDHA-interacting lncRNA GLTC is clinically relevant in PTC
We initiated this study by screening for LDHA-interacting lncRNAs. The profiles obtained by RIP-seq identified a number of candidate LDHA-interacting lncRNAs (Supplementary Fig. 1A). Furthermore, to investigate the oncogenic lncRNAs involved in advanced and aggressive PTC (TNM staging: T ≥ 3, N 1, M 1), we used high-throughput RNA-seq to analyse lncRNA expression in four paired PTC tissues and the corresponding normal thyroid tissues (cohort 1, Fig. 1A, and Supplementary Data 1). Then, using thyroid cancer RNA-seq bioinformatics data from The Cancer Genome Atlas (TCGA), we integrated the LDHA-interacting lncRNA data and lncRNA sequencing data for cohort 1 (Fig. 1B). A total of ten overlapping lncRNAs were finally identified as differentially expressed lncRNAs interacting with LDHA in PTC (Supplementary Table 2). However, only four lncRNAs were overexpressed in the advanced and aggressive PTC datasets. These four lncRNAs (NR2F1-AS1, Lnc-STAT1-2, NEAT1, and Lnc-UNC93B1-1) were selected for further evaluation.
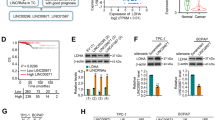
A Flow chart for the selection of candidate lncRNAs by RNA-seq analysis of 4 paired advanced and aggressive PTC tumour tissues and normal thyroid tissues. B A total of 10 overlapping lncRNAs were identified among LDHA-interacting lncRNAs and aberrantly expressed lncRNAs analysed in our four advanced and aggressive PTC tissues (cohort 1) and thyroid cancer data in the TCGA database. C Cell viability, 18F-FDG uptake, and lactate production were measured in TPC-1 cells transfected with negative control siRNA (si-NC) or siRNAs against NR2F1-AS1, Lnc-STAT1-2, NEAT1, or Lnc-UNC93B1-1 (si-NR2F1-AS1, si-Lnc-STAT1-2, si-NEAT1, and si-Lnc-UNC93B1-1), (n = 3). Student’s t test. The bars indicate the means ± SEMs. D The expression of GLTC in 44 paired PTC tissues (cohort 2) was analysed by qRT‒PCR. Paired t-test. E Progression-free survival was analysed and compared in 44 PTC patients with low or high levels of GLTC (cohort 2) by Kaplan–Meier analysis with the log–rank test. F RNAscope ISH analysis of GLTC expression in PTC and matched normal thyroid tissues (cohort 3). Scale bar: 50 μm. G Kaplan–Meier analysis of overall survival based on the GLTC expression status in 82 patients with PTC (cohort 3) was performed, and differences were evaluated by the log-rank test. H The heatmap shows the association of different clinical characteristics with GLTC expression in tumours with high and low GLTC expression in cohort 3. Statistical significance was determined by the χ2 test. I, J Univariate (I) and multivariate (J) analyses of GLTC expression were performed in cohort 3. All bars correspond to 95% confidence intervals. *P < 0.05. **P < 0.01.
For the following experiments, we constructed siRNAs targeting the 4 differentially regulated lncRNAs and transfected them into two human thyroid cancer cell lines to examine cell viability and glycolysis. As shown in Fig. 1C and Supplementary Fig. 1B, cell viability, 18F-FDG uptake, and lactate production were significantly decreased after silencing of NEAT1 or lnc-UNC93B1-1 in PTC cells. Considering the contradictory patterns of NEAT1 expression in our cohort and the TCGA cohort (Supplementary Table 2), we finally selected lnc-UNC93B1-1 for further study. We hereafter refer to this lncRNA as glycolysis-associated lncRNA of thyroid cancer (GLTC). Bioinformatics analysis showed that GLTC is a 627 bp transcript with 4 exons (bp 620–803, 1642–1778, 5730–5875, and 6001–6160) and is located on human chromosome 11q13.2 (Supplementary Fig. 1C). Rapid amplification of 5′ and 3′ cDNA ends (RACE) assays were performed to obtain the full-length sequence of GLTC (Supplementary Fig. 1D right, E right). Sequencing of the PCR products revealed the boundary between the universal anchor primer and the GLTC sequence (Supplementary Fig. 1D left, E left). The GLTC sequence is shown in Supplementary Fig. 1F. To determine whether GLTC is a noncoding transcript, the wild-type (WT) green fluorescent protein (GFP) gene was fused to the C-terminus of the GLTC open reading frame (ORF) to generate the GLTC-ORF-GFP construct. No expression of the GLTC-ORF-GFP fusion protein was observed in transfected cells, indicating that GLTC is a noncoding RNA (Supplementary Fig. 1G).
Consistent with these bioinformatics analysis results, the RT–PCR results verified that GLTC expression was markedly higher in PTC tissues than in adjacent tissues (cohort 2, Fig. 1D, and Supplementary data 2). High GLTC levels were correlated with short progression-free survival times in PTC patients in cohort 2 (Fig. 1E). To further validate the pathological and clinical significance of GLTC expression in PTC, we detected and compared GLTC expression by ISH in an additional 82 pairs of paraffin-embedded PTC and adjacent tissues (cohort 3, Supplementary data 3). In these samples, GLTC was detectable in almost all tumour cells. The expression of GLTC was higher in tumour tissues than in the normal counterparts (Fig. 1F, Supplementary Fig. 1H). A high expression level of GLTC was significantly associated with unfavourable overall survival (Fig. 1G). As shown in Fig. 1H, high GLTC expression was positively correlated with tumour size, the extent of distant metastasis. Univariate and multivariate regression analyses in cohort 3 indicated that GLTC expression was an independent prognostic indicator of PTC (Fig. 1I, J).
To determine the subcellular localisation of GLTC, we evaluated GLTC expression in the nuclear and cytoplasmic fractions of TPC-1 cells by qRT‒PCR. GLTC was localised mainly in the cytoplasm, with some localisation in the nucleus, and this distribution was further confirmed by RNA FISH (Supplementary Fig. 1I, J). Taken together, these results suggest that GLTC is a lncRNA that is highly expressed in PTC.
GLTC promotes cell viability via glycolytic metabolism
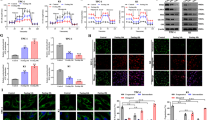
To explore the role of GLTC in the biological behaviours of PTC cells, we first compared the endogenous level of GLTC in four human PTC cell lines with that in the normal thyroid cell line Nthy-ori 3-1 (Supplementary Fig. 2A). We transfected GLTC siRNAs into the PTC cell lines KTC-1 and TPC-1, which express relatively high levels of GLTC (Supplementary Fig. 2B). GLTC knockdown in TPC-1 and KTC-1 cells significantly inhibited cell viability and colony formation (Fig. 2A, B). Furthermore, we determined whether GLTC induces cell viability directly through the aerobic glycolytic pathway. The glycolytic inhibitor 2-deoxy-D-glucose completely blocked the growth-promoting effect of GLTC overexpression in K1 and BCPAP cells (Fig. 2C, Supplementary Fig. 2C), suggesting that aerobic glycolysis might be involved in GLTC-mediated regulation of cell viability. Compared with control cells, cells transfected with GLTC siRNA1/2 exhibited a decreased ECAR and reduced glycolytic activity (Fig. 2D, E). Moreover, lactate production and 18F-FDG uptake were decreased by GLTC knockdown (Fig. 2F, G). The flux of [6-14C]-glucose indicates the activity of glucose metabolism via the mitochondrial pathway, as the release of 14CO2 from [6-14C]-glucose occurs only in the tricarboxylic acid cycle in cancer cells. The proportion of 6-14CO2 in the total CO2 released was increased in KTC-1 and TPC-1 cells with GLTC silencing (Fig. 2H). As expected, these effects were reversed in GLTC-overexpressing cells (Supplementary Fig. 2D–H).
A Cell viability was determined in TPC-1 and KTC-1 cells transfected with the negative control siRNA (si-NC) or GLTC siRNAs (si-GLTC#1, si-GLTC#2), (n = 3). B Colony formation assays of TPC-1 and KTC-1 cells transfected with the negative control siRNA (si-NC) or GLTC siRNAs (si-GLTC#1, si-GLTC#2), (n = 3). C K1 and BCPAP cells transfected with empty vector (vector) or the pCDNA-GLTC plasmid (pCDNA-GLTC) were treated or not treated with 2-deoxy-D-glucose (2-DG, 2.5 mM), (n = 3). Cell viability was then analysed. D The ECAR in TPC-1 and KTC-1 cells transfected with the negative control siRNA (si-NC) or GLTC siRNAs (si-GLTC#1, si-GLTC#2) was measured using an XF Extracellular Flux Analyser, (n = 3). E Statistical analysis of the effects of GLTC knockdown on glycolytic activity, (n = 3). Glycolytic parameters were obtained from (D) as follows: glycolysis = (ECAR after glucose addition – basal ECAR); glycolytic capacity = (ECAR after oligomycin addition – basal ECAR); and glycolytic reserve = (glycolytic capacity – glycolysis). F–H Lactate level (F), 18F-FDG uptake (G), and 6-14CO2 production from [6-14C]-glucose (H) in TPC-1 and KTC-1 cells transfected with the negative control siRNA (si-NC) or GLTC siRNAs (si-GLTC#1, si-GLTC#2), (n = 3). I Nude mice were injected with 1 × 107 TPC-1 cells stably expressing the negative control shRNA (sh-NC) or GLTC shRNA (sh-GLTC), (n = 5 mice per group). Representative images of the in vivo glucose uptake assay by 18F-FDG micro-PET scanning (left panel). Comparison of the maximum standard uptake value (SUVmax) between xenograft tumours derived from sh-NC and sh-GLTC cells (right panel). J Image of tumours isolated from nude mice. K Tumour weights were measured in the sh-NC and sh-GLTC groups, (n = 5 mice per group). L Left panel: Representative images of haematoxylin and eosin (H/E) staining and Ki67 staining in tumour sections from the sh-NC and sh-GLTC groups (scale bar, 50 mm), (n = 5 mice per group). Right panel: semiquantification of the Ki67 index in tumour samples. The Ki67 index was calculated by determining the percentage of Ki67-positive cells. M Lactate production was measured in lysates derived from xenografts in the sh-NC and sh-GLTC groups, (n = 5 mice per group). The P values in (A–C) and (E–H) were determined by one-way ANOVA. The P values in (I) and (K–M) were determined by two-tailed unpaired Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. NS not significant.
Then, we investigated the roles of GLTC in the glucose metabolic phenotype and tumorigenesis in vivo. We performed 18F-FDG micro-PET imaging and found that compared with tumours derived from control cells, tumours derived from sh-GLTC TPC-1 cells exhibited decreased glucose uptake (Fig. 2I and Supplementary Fig. 2I). GLTC knockdown significantly reduced xenograft tumour growth, the level of the cell viability marker Ki67, and lactate production (Fig. 2J–M). Collectively, these results suggest that GLTC promotes glycolytic metabolism in PTC cells and the growth of PTC tumours.
GLTC interacts with LDHA and regulates LDHA activity
We further explored the mechanistic role of the LDHA-interacting lncRNA GLTC in metabolic remodelling in PTC cells. The GLTC pulldown assay followed by mass spectrometry showed that LDHA is a candidate RNA-binding protein (Fig. 3A). Subsequent western blot analysis demonstrated that LDHA bound specifically to the sense GLTC probe but not to the antisense GLTC probe (Fig. 3B). To identify the LDHA-interacting region of GLTC, we constructed four different deletion fragments of GLTC (F1: full-length sense GLTC, F2: full-length antisense GLTC, Δ1: (1–250 bp), Δ2: (1–500 bp), Δ3: (251–627 bp). Subsequently, a GLTC fragment RNA pulldown assay followed by western blotting showed that the fragment of GLTC containing nucleotides 1–250 specifically mediated the interaction with LDHA (Fig. 3C). Further validation of binding showed specific enrichment of GLTC in the coprecipitated LDHA immunocomplex (Fig. 3D). We constructed expression plasmids containing HA-tagged full-length LDHA, ΔN, ΔC, ΔLDHA 151–208, ΔLDHA 209–268, and ΔLDHA 269–332 and transfected them into TPC-1 cells (Fig. 3E). Real-time PCR analysis following RIP confirmed that the region of LDHA containing amino acids 151–208 was essential for the binding of this protein to GLTC (Fig. 3F).
A Proteins retrieved in the GLTC pulldown assay were analysed by mass spectrometry. Proteome-wide accurate quantification and significance are presented. B Western blot analysis of LDHA protein pulled down by the antisense and sense GLTC probes in the pulldown assay, (n = 3). C Western blot analysis of LDHA in samples pulled down by the full-length sense GLTC probe (Sense), antisense GLTC probe (Antisense), and truncated forms of GLTC (Δ1: 1–250, Δ2: 1–500, Δ3: 251–627), (n = 3). D TPC-1 cell lysates were incubated with magnetic beads conjugated to an anti- LDHA antibody or the corresponding negative control (IgG) at 4 °C for 6 h, (n = 3). RNA was extracted and subjected to qRT–PCR using GLTC-specific primers. A cell lysate sample not subjected to immunoprecipitation was used as the qRT–PCR control (input). E Expression plasmids containing HA-tagged full-length LDHA and truncated forms of LDHA (ΔN: ΔLDHA 1–150, ΔC: ΔLDHA 151–332, ΔLDHA 151–208, ΔLDHA 209–268, ΔLDHA 269–332) were separately transfected into TPC-1 cells for 48 h. Western blot analysis was performed to evaluate the expression of WT LDHA and the LDHA truncations in TPC-1 cells, (n = 3). F qRT–PCR was performed to evaluate the binding efficiency between LDHA (LDHA and all truncated forms of LDHA) proteins and GLTC in the RIP assay, (n = 3). G–I TPC-1 cells were transfected with the negative control siRNA (si-NC) or a GLTC siRNA (si-GLTC#1, si-GLTC#2) for 48 h, (n = 3). K1 cells were transfected with pCDNA-GLTC or empty vector control for 48 h, (n = 3). The level of LDHA mRNA was measured by qRT–PCR (G). The LDHA protein level was examined by western blotting (H). HA-LDHA was immunoprecipitated from the lysates of all transfected cells, and LDHA activity was assayed. LDHA activity was normalised to the protein level (I). J LDHA activity was measured in K1 cells transfected with the pCDNA-GLTC, pCDNA-GLTC-Δ3 or empty vector control and the HA-LDHA plasmid, (n = 3). HA-LDHA was immunoprecipitated, and LDHA activity was assayed. LDHA activity was normalised to the protein level. K–M Cell viability (K), ECAR (L), and lactate production assays (M) were performed in K1 cells transfected with the pCDNA-GLTC, pCDNA-GLTC-Δ3 or empty vector control, (n = 3). Statistical analysis of the mean ECAR values (L) in K1 cells transfected with the GLTC-WT or GLTC-Δ3 overexpression plasmid (or empty vector) are shown. Glycolytic parameters were calculated as follows: glycolysis = (ECAR after glucose addition – basal ECAR); glycolytic capacity = (ECAR after oligomycin addition – basal ECAR); and glycolytic reserve = (glycolytic capacity – glycolysis). The P values in (F, G) (left panel), (I) (left panel), and (J–M) were determined by one-way ANOVA. The P values in (D, G) (right panel), and (I) (right panel) were determined by two-tailed unpaired Student’s t test. *P < 0.05, **P < 0.01. NS not significant.
We next investigated whether overexpression of GLTC regulates LDHA expression or function in PTC cells. GLTC knockdown had no effect on the mRNA or protein level of LDHA (Fig. 3G, H; Supplementary Fig. 3A, B) but resulted in a decrease in LDHA activity in PTC cells (Fig. 3I, Supplementary Fig. 3C). Moreover, expression of WT GLTC but not the Δ3 fragment of GLTC resulted in increased LDHA activity, cell viability and glycolytic metabolism (Fig. 3J–M).
GLTC increases LDHA succinylation at K155
Recent studies have shown that posttranslational modifications (PTMs), as dynamic modification processes, might affect the activity of many metabolic enzymes [15]. Indeed, GLTC overexpression increased LDHA succinylation in K1 cells (Fig. 4A). In support of this observation, GLTC knockdown decreased LDHA succinylation (Fig. 4B). The acetylation, malonylation and glutarylation of LDHA remained largely unaffected by GLTC (Fig. 4C).
A, B K1 cells were cotransfected with HA-LDHA and pCDNA-GLTC (or the empty vector control) for 48 h (A), (n = 3). TPC-1 cells were cotransfected with HA-LDHA and si-GLTC#1 (or si-NC as the negative control siRNA) for 48 h (B), (n = 3). Cell lysates were immunoprecipitated with HA-LDHA affinity gel. LDHA succinylation and protein levels were analysed by immunoblotting using the indicated antibodies. C K1 cells were transfected with HA-LDHA and pCDNA-GLTC (or the empty vector control) for 48 h, (n = 3). Cell lysates were immunoprecipitated with HA-LDHA affinity gel. LDHA acetylation (Ac), malonylation (Mal), and glutarylation (Glu) were analysed by immunoblotting using the indicated antibodies. D Identification of succinylated LDHA peptides by mass spectrometry in K1 cells transfected with the empty vector control (vector) or pCDNA-GLTC plasmid (GLTC). E LC‒MS/MS spectrum of the K155-succinylated peptide of LDHA. F Alignment of protein sequences surrounding K155 of LDHA from different organisms. Xl: Xenopus laevis, frog; Mm: Mus musculus, mouse; Ss: Sus scrofa, pig; Rn: Rattus norvegicus, Norway rat; Hs: Homo sapiens, human. G HA-LDHAWT, HA-LDHAK155R or HA-LDHAK155E proteins were expressed in TPC-1 cells and purified by immunoprecipitation, (n = 3). LDHA enzymatic activity was measured and normalised to the protein level. H, I Western blot analysis of K155-succinylated LDHA and endogenous LDHA levels in TPC-1 cells transfected with si-GLTC#1/2 (or si-NC as the negative control siRNA) and in K1 cells transfected with pCDNA-GLTC (or the empty vector control), (n = 3). J, K TPC-1 cells were cotransfected with si-GLTC#1 (or si-NC as the negative control siRNA) and HA-LDHAWT, HA-LDHAK155R or HA-LDHAK155E for 48 h (J), (n = 3). K1 cells were cotransfected with pCDNA-GLTC (or the empty vector control) and HA-LDHAWT, HA-LDHAK155R or HA-LDHAK155E for 48 h (K), (n = 3). LDHA was immunoprecipitated. Succinylation of K155 was evaluated by western blotting, and LDHA activity was assayed. The P value in (G) was determined by one-way ANOVA. The P values in (J and K) were determined by two-tailed unpaired Student’s t test. *P < 0.05, **P < 0.01. NS not significant.
To verify the site of LDHA succinylation regulated by GLTC, we analysed succinylated peptides from K1 cells with or without GLTC transfection by LC‒MS/MS. LDHA was succinylated at K155 in GLTC-transfected cells, where the level of LDHA-K155 succinylation was 2.822-fold higher than that in the vector control cells (Fig. 4D, E). K155 is evolutionarily conserved from Xenopus laevis to mammals (Fig. 4F).
Then, we generated a single succinylation mimetic (K-to-glutamine, E) mutant (LDHAK155E) and succinylation-null (K-to-arginine, R) mutant (LDHAK155R) and examined their enzymatic activity. The activity of LDHAK155R was only 50% of that of WT LDHA, whereas the LDHAK155E mutation resulted in a 50% increase in LDHA activity in TPC-1 cells (Fig. 4G). Furthermore, we generated an antibody specifically recognising K155-succinylated LDHA. Western blot analysis using this antibody showed that the level of K155-succinylated endogenous LDHA was reduced by suppression of GLTC expression, whereas GLTC overexpression enhanced LDHA K155 succinylation in PTC cells (Fig. 4H, I). GLTC regulated the activity of WT LDHA but not the LDHAK155R and LDHAK155E mutants (Fig. 4J, K), indicating that GLTC stimulates LDHA activity mostly via succinylation at K155.
The GLTC/LDHA interaction disrupts SIRT5-mediated modification of LDHA in PTC
SIRT5 regulates the activity of many metabolic enzymes via PTMs. In a previous study, an interaction was found between SIRT5 and LDHA in the prostate cancer cell line PC-3 [21]. The SIRT5–LDHA interaction was confirmed by endogenous and exogenous co-IP in TPC-1 cells (Fig. 5A, B; Supplementary Fig. 4A, B). Then, GLTC and GLTC-Δ3 were transfected into GLTC knockdown TPC-1 cells, and the LDHA-SIRT5 interaction was evaluated. The LDHA/SIRT5 interaction was partially weakened by GLTC overexpression but not by GLTC-Δ3 overexpression (Fig. 5C). We next explored whether SIRT5 is involved in modulating the GLTC-induced succinylation of LDHA and found that SIRT5 induced LDHA-K155 desuccinylation in PTC cells (Fig. 5D, E). More importantly, overexpression of GLTC abolished SIRT5-induced LDHA-K155 desuccinylation, while knockdown of GLTC attenuated the succinylation of LDHA-K155 mediated by SIRT5 suppression (Fig. 5D, E). LDHA is a crucial glycolytic enzyme that catalyses the reduction of pyruvate to lactate with the conversion of NADH and H+ to NAD+ [22] (Supplementary Fig. 4C), and we investigated whether SIRT5 participates in GLTC-regulated LDHA enzymatic activity and modulates the cellular lactate level and NAD+/NADH ratio. As shown in Fig. 5F–K, forced expression of SIRT5 diminished the GLTC-induced increases in LDHA activity, lactate production and the NAD+/NADH ratio, whereas knockdown of SIRT5 restored the effect of si-GLTC on decreasing LDHA activity, lactate production, and the NAD+/NADH ratio in PTC cells. Collectively, our data demonstrate that GLTC enhances LDHA-K155 succinylation by disrupting the SIRT5-directed interaction with LDHA, which consequently enhances LDHA activity in PTC cells.
A Lysates of TPC-1 cells were immunoprecipitated with control IgG or an anti-LDHA antibody, (n = 3). LDHA and SIRT5 expression levels were determined by western blotting. B TPC-1 cells were transfected with HA-LDHA for 48 h, (n = 3). Immunoprecipitation was performed using control IgG or an anti-HA antibody, and western blotting was then performed with an anti-SIRT5 or anti-HA antibody. C SIRT5/LDHA interactions were evaluated in four groups of transfected TPC-1 cells (sh-NC + vector; sh-GLTC + vector; sh-GLTC + GLTC; sh-GLTC + GLTC-Δ3) by IP and western blotting, (n = 3). D K1 cells were cotransfected with pCDNA-GLTC (or the empty vector control) and Flag-SIRT5 (or the empty vector control) for 48 h, (n = 3). LDHA-K155 succinylation was evaluated by western blotting. E TPC-1 cells were cotransfected with si-GLTC#1 (or si-NC as the negative control siRNA) and si-SIRT5 (or si-NC as the negative control siRNA) for 48 h. LDHA-K155 succinylation was evaluated by western blotting, (n = 3). F, G LDHA protein was immunoprecipitated from the cells used in (D and E). LDHA activity was assayed and normalised to the protein level, (n = 3). H–K Lactate production (H, I) and the NAD + /NADH ratio (J, K) in the cells used in (D and E) were determined, (n = 3). The P values in (F–K) were determined by one-way ANOVA. *P < 0.05, **P < 0.01.
LDHA succinylation at K155 contributes to GLTC-induced promotion of glycolysis and tumour progression
We next hypothesised that LDHA-K155 succinylation mediates the biological function of GLTC in PTC. To test this hypothesis, we generated TPC-1 LDHA-KO cells and transfected them with the HA-LDHAWT, HA-LDHAK155R, and HA-LDHAK155E plasmids. Intriguingly, the K155R mutant decreased but the K155E mutant increased cell viability, the ECAR, 18F-FDG uptake, and lactate production compared with those in the LDHA WT group in TPC-1 cells (Fig. 6A–D and Supplementary Fig. 5A). Moreover, when GLTC was knocked down in these cells, WT LDHA exhibited an effect similar to that of the K155R mutant on cell viability, while the K155E mutant still promoted cell viability, the ECAR, 18F-FDG uptake, and lactate production (Fig. 6A–D).
A–D LDHA KO TPC-1 cells were cotransfected with si-GLTC#1 (or si-NC as the negative control) and HA-LDHAWT, HA-LDHAK155R or HA-LDHAK155E. Cell viability (A), the ECAR (B), 18F-FDG uptake (C), and lactate production (D) were measured, (n = 3). E–G A total of 1.0 × 107 LDHA-KO K1 cells stably coexpressing the GLTC plasmid (or the empty vector control) and HA-LDHAWT, HA-LDHAK155R or HA-LDHAK155E were injected subcutaneously into the dorsal flanks of nude mice, (n = 5 mice per group). Image of tumours isolated from nude mice (E). The weight of tumours when mice were sacrificed (F). LDHA protein and LDHA-K155 succinylation levels were measured by western blotting using whole-cell lysates from xenograft tumour tissues from each group (G). H Representative RNAscope ISH analysis of GLTC expression and IHC analysis of LDHA-K155 succinylation in PTC samples from cohort 3, (n = 82). Scale bars, 50 μm in each image. I Statistical analysis of thyroid cancer tissues in cohort 3 under different staining conditions. Chi-square test. The P values in (A–D) and (F) were determined by one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001. NS not significant.
After the cell viability and glycolytic advantages afforded by GLTC-mediated LDHA succinylation were verified in vitro, we performed xenograft studies in vivo. LDHA-KO K1 cells stably expressing the HA-LDHAWT, HA-LDHAK155R, or HA-LDHAK155E and the GLTC or control overexpression plasmids were inoculated into nude mice, and the K155R mutant slowed but the K155E mutant accelerated tumour growth compared with that observed with WT LDHA (Fig. 6E, F and Supplementary Fig. 5B). In GLTC-overexpressing PTC xenograft tumours, the K155E mutant exhibited an effect similar to that of WT LDHA on tumour growth, while the K155R mutant still significantly blocked tumour growth. The regulatory effect of GLTC on K155 succinylation of LDHA was confirmed by western blot analysis in xenograft samples from different groups (Fig. 6G). More importantly, when GLTC was overexpressed, tumours expressing HA-LDHAWT exhibited lactate production and NAD+/NADH ratios similar to those of tumours expressing the K155E mutant, while the corresponding values in tumours expressing the K155R mutant remained low (Supplementary Fig. 5C, D).
We next performed ISH and IHC staining on PTC patient tissues from cohort 3. As expected, samples with GLTC expression above the median value showed strong staining for K155-succinylated LDHA, and samples with GLTC expression below the median value exhibited low levels of LDHA-K155 succinylation (Fig. 6H, I, Supplementary Fig. 5E). However, the SIRT5 expression levels in extracts from xenograft tumour tissue samples and in PTC patient tissues did not fluctuate in response to changes in GLTC expression, eliminating the possibility that SIRT5 is affected by GLTC (Supplementary Fig. 5F, G). Collectively, these data indicate that GLTC-mediated regulation of glycolytic metabolism and tumour progression depend on K155 succinylation of LDHA in PTC.
GLTC enhances 131I resistance in PTC cells via an LDHA-K155 succinylation-mediated increase in cellular NAD+
RAI is the most successful treatment for postoperative PTC patients. However, some PTCs do not respond to RAI and become refractory to RAI, with an unfavourable prognosis. RAI resistance has become a major problem for anticancer therapy. Thus, we generated a 131I-resistant thyroid cancer TPC-1 cell line (TPC-1/IR) by continuous exposure to a sublethal 131I concentration until resistance to 131I developed (Fig. 7A). To determine a possible role of GLTC in 131I resistance, we studied the expression levels of GLTC and its target, K155-succinylated LDHA, as well as LDHA activity in TPC-1 cells in response to 131I treatment. As shown in Fig. 7B–D, GLTC expression, LDHA-K155 succinylation and LDHA activity were increased in TPC-1 cells in a 131I concentration-dependent manner. As expected, GLTC expression, LDHA-K155 succinylation, and LDHA activity were increased more in TPC-1/IR cells than in TPC-1 cells (Fig. 7E, F). To investigate whether GLTC and LDHA confer 131I resistance on PTC cells, we analysed the effects of GLTC and LDHA knockdown on the survival rates of TPC-1/IR cells following 131I treatment. Knockdown of GLTC and LDHA, respectively, resulted in a decrease in cell survival rate of TPC-1/IR cells (Fig. 7G). TPC-1 cells stably expressing LDHAK155R also had decreased resistance to 131I compared with those stably expressing LDHAWT or LDHAK155E, suggesting that succinylation of LDHA at K155 plays a crucial role in 131I resistance in PTC cells (Fig. 7H). Because 131I-resistant cells had increased GLTC expression, we explored whether GLTC knockdown can sensitise 131I-resistant cells to 131I. Treatment with the combination of 131I and si-GLTC synergistically inhibited the growth of TPC-1/IR cells (Fig. 7I).
A TPC-1 and TPC-1/IR cells were treated for 2 days with increasing concentrations of 131I, and their survival rates were determined by cell counting, (n = 3). B–D TPC-1 cells were treated with 0, 0.4, 0.8, or 1.6 mCi 131I for 24 h, (n = 3). qPCR analysis of GLTC expression (B) and western blot analysis of LDHA-K155 succinylation (C). Endogenous LDHA was immunoprecipitated, and LDHA activity was assayed (D). LDHA activity was normalised to the protein level. E qPCR analysis of GLTC expression (left panel) and western blot analysis of LDHA K155 succinylation (right panel) in TPC-1 and TPC-1/IR cells, (n = 3). F Endogenous LDHA was immunoprecipitated from TPC-1 and TPC-1/IR cells, (n = 3). LDHA activity was assayed. LDHA activity was normalised to the protein level. G TPC-1/IR cells transfected with si-GLTC#1, si-LDHA or the negative control siRNA (si-NC) were treated with 2.8 mCi 131I for 2 days, (n = 3). Cell survival rates were determined by cell counting. H LDHA-KO TPC-1 cells expressing HA-LDHAWT, HA-LDHAK155R or HA-LDHAK155E were treated with 0.8 mCi 131I for 2 days, (n = 3). Cell survival rates were determined by cell counting. I TPC-1/IR cells were transfected with the negative control siRNA (si-NC) or si-GLTC#1 for 48 h and were then treated with PBS (control) or 2.8 mCi 131I, (n = 3). Cell survival rates were determined by cell counting. J TPC-1 cells were transfected with si-GLTC#1 (or si-NC as the negative control siRNA) for 48 h, (n = 3). The cells were cocultured with 0.8 mCi 131I or 10 mM NAD+ (mock treatment with PBS). The p-H2AX level was measured by western blotting. K TPC-1/IR cells expressing sh-NC or sh-GLTC were injected into nude mice. Fourteen days after tumour cell implantation, the mice were randomly assigned to a group receiving 131I treatment (2.8 mCi in 100 μL intratumorally) or a negative control group (mock treatment with saline). (n = 5 mice per group). L After 3 weeks of treatment, the mice were sacrificed, and the tumours were weighed, (n = 5 mice per group). M Left panel: H/E staining and Ki67 staining of tumour samples (scale bar, 50 mm). Right panel: semiquantification of the Ki67 index in tumour samples, (n = 5 mice per group). The Ki67 index was calculated by determining the percentage of Ki67-positive cells. The P values in (B, D, H, I, L, and M) were determined by one-way ANOVA. The P values in (E–G) were determined by two-tailed unpaired Student’s t test. *P < 0.05, **P < 0.01, *** P < 0.001. NS not significant.
A previous study showed that NAD+ can prevent radiation-induced dsDNA damage and inhibit the dsDNA-induced cell death pathway [23]. NAD+ is the product of the reduction reaction catalysed by LDHA [24], and our results demonstrated that GLTC-mediated succinylation of LDHA increased the NAD+/NADH ratio. We thus hypothesised that NAD+ might be involved in 131I resistance in TPC-1 cells via 131I radiation-induced dsDNA damage and the cell death pathway. Indeed, GLTC knockdown induced a marked increase in the level of γ-H2AX, a marker of dsDNA damage, in TPC-1 cells after 131I treatment (Fig. 7J).
To further confirm these results in vivo, we treated BALB/c nude mice bearing subcutaneous xenograft tumours derived from TPC-1/IR cells stably expressing sh-GLTC or sh-NC with 2.8 mCi 131I. Compared with the control and single-agent treatments, combination treatment with GLTC knockdown and 131I more efficiently inhibited TPC-1/IR tumour growth (Fig. 7K, L). Ki67 staining showed that combination treatment with sh-GLTC and 131I resulted in a significant inhibitory effect on the cell viability of TPC-1/IR cells (Fig. 7M). Collectively, these results further suggest that GLTC-mediated LDHA succinylation contributes to 131I resistance.
Discussion
Many studies have indicated that lncRNAs are involved in the regulation of the Warburg effect [25, 26], but the potential involvement of lncRNAs is poorly defined in human PTC. Here, we found that the lncRNA GLTC is an oncogenic lncRNA in PTC. In cultured PTC cells and xenograft mouse models, GLTC promoted cell growth and glycolysis. The data consistently indicated that high GLTC expression is a crucial factor controlling the aggressiveness of human PTC.
Cytoplasmic lncRNAs may exert their biological effects by reacting with RNA-binding proteins, which may in turn regulate protein activity or stability in various ways [27, 28]. Our RNA pulldown and subsequent mass spectrometry experiments demonstrated that cytoplasmic GLTC directly interacts with the LDHA protein and further increases the enzymatic activity of LDHA, which then activates glycolytic metabolism in tumour cells. A few metabolic enzymes have been established as novel regulators of thyroid cancer growth and metabolism [29, 30]. LDHA is an oncogenic metabolic enzyme that initiates a phenotypic shift from mitochondrial respiration to glycolysis [31, 32]. Based on data from the TCGA database, LDHA has been determined to be a potential novel diagnostic and therapeutic target in PTC [33]. Elevated LDHA mRNA and protein levels significantly impact PTC progression [34]. Noncoding RNAs, such as microRNAs and lncRNAs, have also been reported to play critical roles in targeting LDHA. For example, miR-33b-5p directly binds to the LDHA 3′UTR to regulate LDHA expression [35]. LINC00671 is reported to activate LDHA expression largely through regulation of STAT3 transcription [36]. Our results indicated that nucleotides 1–250 of GLTC and amino acids 151-208 of the LDHA protein are required for their interaction. The Δ3 fragment of GLTC (with genetic deletion of nucleotides 1–250) lost the ability to bind to LDHA and disrupted the enzymatic activity of LDHA, leading to inhibition of glycolysis and cell viability in PTC cells. Thus, GLTC might be a crucial regulator of PTC progression via a direct interaction with LDHA.
We explored the mechanisms by which GLTC contributes to LDHA activity enhancement and found that PTM of LDHA might be involved. PTMs regulate the function of multiple metabolic enzymes and, consequently, cellular biochemical metabolism [15, 37, 38]. LDHA expression and function can also be regulated posttranslationally. Succinylation of LDHA at K222 inhibits its lysosomal degradation and causes LDHA accumulation in the cytoplasm, leading to increased glycolysis and cell viability in gastric carcinoma cells [39]. LDHA-K5 acetylation inhibits LDHA activity and promotes its degradation [40]. In addition, phosphorylation of LDHA at Y10 increases its activity and confers anti-anoikis, proinvasive, and prometastatic potential on cancer cells [41]. However, the status of LDHA succinylation in PTC cells and the upstream factors are still unclear. We found that succinylation of LDHA was closely correlated with GLTC expression. Moreover, we demonstrated that GLTC increased the succinylation of LDHA at K155 to elevate LDHA activity, leading to enhancement of glucose uptake, lactate accumulation, and an increase in the NAD+/NADH ratio. Our results reveal a link between oncogenic GLTC and the tumour glycolytic enzyme LDHA whereby GLTC controls the metabolic switch by regulating succinylation-mediated enzymatic activity in tumour cells.
SIRT5, a member of the sirtuin family, has been demonstrated to regulate metabolic enzymes via PTM and control several cellular metabolic pathways, including the tricarboxylic acid cycle and glycolysis [42, 43]. This study revealed that SIRT5 is responsible for the regulation of LDHA desuccinylation by directly binding to the LDHA protein in PTC cells. Collectively, these findings further delineate the detailed mechanism underlying GLTC-mediated LDHA activity increase and cancer metabolic remodelling.
RAI is considered a gold standard in the treatment of PTC patients [44]. However, two-thirds of patients with metastatic PTC become refractory to RAI. RAI resistance remains the main cause of death during PTC therapy, with a 10-year survival rate of affected patients as low as 10% [45]. Although studies have investigated RAI refractoriness in PTC cells [46], the molecular mechanisms that mediate this phenomenon remain unknown. Previous studies showed that high 18F-FDG uptake in metastatic PTC indicated a poor response to RAI therapy [47]. This conclusion was consistent with our experimental results indicating that succinylation of LDHA by GTLC enhanced glycolysis and subsequent NAD+ production, which finally contributed to the resistance of PTC cells to RAI.
LncRNAs are emerging as key molecules with the potential to confer many molecular phenotypic characteristics of PTC and to serve as novel targets in PTC [48]. Our study showed that GLTC is highly expressed in PTC and that high expression of GLTC is closely linked to unfavourable outcomes, suggesting that GLTC is a diagnostic and prognostic marker and therapeutic target in PTC.
In conclusion, we found that GLTC plays an important role in promoting cell viability as well as glycolytic pathway activity by enhancing LDHA enzymatic activity. Intriguingly, this is the first lncRNA proven to directly interact with and regulate LDHA. GLTC facilitates the succinylation-dependent activation of LDHA by competitive binding to LDHA with SIRT5, a PTM enzyme mediating LDHA desuccinylation at K155 (Fig. 8). Furthermore, enhancement of LDHA activity mediated by GLTC promotes RAI resistance in vitro and in vivo. A high GLTC expression level is correlated with poor overall survival in PTC. These data provide the rationale for further use of the GLTC–LDHA pathway as a potential target for therapeutic intervention in PTC.
Data availability
All data generated or analysed during this study are included in this published paper and its supplementary information files. RNA-Seq data have been deposited in NCBI SRA database (PRJNA857849).
References
Megwalu UC, Moon PK. Thyroid cancer incidence and mortality trends in the United States: 2000-2018. Thyroid. 2022;32:560–70.
Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in thyroid cancer incidence and mortality in the United States, 1974-2013. Jama. 2017;317:1338–48.
Luo H, Xia X, Kim GD, Liu Y, Xue Z, Zhang L, et al. Characterizing dedifferentiation of thyroid cancer by integrated analysis. Sci Adv. 2021;7:eabf3657.
Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381:1058–69.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Robbins RJ, Wan Q, Grewal RK, Reibke R, Gonen M, Strauss HW, et al. Real-time prognosis for metastatic thyroid carcinoma based on 2-[18F] fluoro-2-deoxy-D-glucose-positron emission tomography scanning. J Clin Endocrinol Metab. 2006;91:498–505.
Wang W, Larson SM, Tuttle RM, Kalaigian H, Kolbert K, Sonenberg M, et al. Resistance of [18f]-fluorodeoxyglucose-avid metastatic thyroid cancer lesions to treatment with high-dose radioactive iodine. Thyroid. 2001;11:1169–75.
Augoff K, Hryniewicz-Jankowska A, Tabola R. Lactate dehydrogenase 5: an old friend and a new hope in the war on cancer. Cancer Lett. 2015;358:1–7.
de la Cruz-López KG, Castro-Muñoz LJ, Reyes-Hernández DO, García-Carrancá A, Manzo-Merino J. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol. 2019;9:1143.
Brooks GA. The science and translation of lactate shuttle theory. Cell Metab. 2018;27:757–85.
Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18.
Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer. 2019;18:135.
Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. 2019;12:3.
Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17:141.
Shi L, Yan H, An S, Shen M, Jia W, Zhang R, et al. SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol Oncol. 2019;13:358–75.
Shen M, Zhao X, Zhao L, Shi L, An S, Huang G, et al. Met is involved in TIGAR-regulated metastasis of non-small-cell lung cancer. Mol Cancer. 2018;17:88.
Shen M, Zhang R, Jia W, Zhu Z, Zhao X, Zhao L, et al. Nuclear scaffold protein p54(nrb)/NONO facilitates the hypoxia-enhanced progression of hepatocellular carcinoma. Oncogene. 2021;40:4167–83.
Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, et al. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93:4331–41.
Kurebayashi J, Tanaka K, Otsuki T, Moriya T, Kunisue H, Uno M, et al. All-trans-retinoic acid modulates expression levels of thyroglobulin and cytokines in a new human poorly differentiated papillary thyroid carcinoma cell line, KTC-1. J Clin Endocrinol Metab. 2000;85:2889–96.
Li L, Liang Y, Kang L, Liu Y, Gao S, Chen S, et al. Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer Cell. 2018;33:368–85. Mar
Kwon OK, Bang IH, Choi SY, Jeon JM, Na AY, Gao Y, et al. SIRT5 is the desuccinylase of LDHA as novel cancer metastatic stimulator in aggressive prostate cancer. Genom Proteom Bioinform. 2022;S1672-0229(22)00018-3.
Quistorff B, Grunnet N. High brain lactate is not caused by a shift in the lactate dehydrogenase A/B ratio. Proc Natl Acad Sci U S A. 2011;108:E21. author reply E2
Sheng C, Chen H, Wang B, Liu T, Hong Y, Shao J, et al. NAD(+) administration significantly attenuates synchrotron radiation X-ray-induced DNA damage and structural alterations of rodent testes. Int J Physiol Pathophysiol Pharmacol. 2012;4:1–9.
Pathria G, Scott DA, Feng Y, Sang Lee J, Fujita Y, Zhang G, et al. Targeting the Warburg effect via LDHA inhibition engages ATF4 signaling for cancer cell survival. Embo J. 2018;37:e99735.
Shankaraiah RC, Veronese A, Sabbioni S, Negrini M. Non-coding RNAs in the reprogramming of glucose metabolism in cancer. Cancer Lett. 2018;419:167–74.
Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21:498–510.
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–3.
Li Z, Zhang J, Liu X, Li S, Wang Q, Di C, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. 2018;9:1572.
Thakur S, Daley B, Gaskins K, Vasko VV, Boufraqech M, Patel D, et al. Metformin targets mitochondrial glycerophosphate dehydrogenase to control rate of oxidative phosphorylation and growth of thyroid cancer in vitro and in vivo. Clin Cancer Res. 2018;24:4030–43.
Huang J, Gao W, Liu H, Yin G, Duan H, Huang Z, et al. Up-regulated ANP32E promotes the thyroid carcinoma cell proliferation and migration via activating AKT/mTOR/HK2-mediated glycolysis. Gene. 2020;750:144681.
Icard P, Loi M, Wu Z, Ginguay A, Lincet H, Robin E, et al. Metabolic strategies for inhibiting cancer development. Adv Nutr. 2021;12:1461–80.
Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152.
Ban EJ, Kim D, Kim JK, Kang SW, Lee J, Jeong JJ, et al. Lactate dehydrogenase A as a potential new biomarker for thyroid cancer. Endocrinol Metab (Seoul). 2021;36:96–105.
Hou X, Shi X, Zhang W, Li D, Hu L, Yang J, et al. LDHA induces EMT gene transcription and regulates autophagy to promote the metastasis and tumorigenesis of papillary thyroid carcinoma. Cell Death Dis. 2021;12:347.
Wang YY, Chen C. lncRNA-DANCR promotes taxol resistance of prostate cancer cells through modulating the miR-33b-5p-LDHA axis. Dis Markers. 2022;2022:9516774.
Huo N, Cong R, Sun ZJ, Li WC, Zhu X, Xue CY, et al. STAT3/LINC00671 axis regulates papillary thyroid tumor growth and metastasis via LDHA-mediated glycolysis. Cell Death Dis. 2021;12:799.
Shangguan X, He J, Ma Z, Zhang W, Ji Y, Shen K, et al. SUMOylation controls the binding of hexokinase 2 to mitochondria and protects against prostate cancer tumorigenesis. Nat Commun. 2021;12:1812.
Coassolo S, Davidson G, Negroni L, Gambi G, Daujat S, Romier C, et al. Citrullination of pyruvate kinase M2 by PADI1 and PADI3 regulates glycolysis and cancer cell proliferation. Nat Commun. 2021;12:1718.
Li X, Zhang C, Zhao T, Su Z, Li M, Hu J, et al. Lysine-222 succinylation reduces lysosomal degradation of lactate dehydrogenase a and is increased in gastric cancer. J Exp Clin Cancer Res. 2020;39:172.
Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P, et al. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 2013;23:464–76.
Jin L, Chun J, Pan C, Alesi GN, Li D, Magliocca KR, et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene. 2017;36:3797–806.
Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–8.
Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–30.
Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016;26:1–133.
Durante C, Haddy N, Baudin E, Leboulleux S, Hartl D, Travagli JP, et al. Long-term outcome of 444 patients with distant metastases from papillary and follicular thyroid carcinoma: benefits and limits of radioiodine therapy. J Clin Endocrinol Metab. 2006;91:2892–9.
Oh JM, Ahn BC. Molecular mechanisms of radioactive iodine refractoriness in differentiated thyroid cancer: Impaired sodium iodide symporter (NIS) expression owing to altered signaling pathway activity and intracellular localization of NIS. Theranostics. 2021;11:6251–77.
Kang SY, Bang JI, Kang KW, Lee HY, Chung JK. FDG PET/CT for the early prediction of RAI therapy response in patients with metastatic differentiated thyroid carcinoma. PLoS ONE. 2019;14:e0218416.
Lin RX, Yang SL, Jia Y, Wu JC, Xu Z, Zhang H. Epigenetic regulation of papillary thyroid carcinoma by long non-coding RNAs. Semin Cancer Biol. 2022;83:253–60.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82001865), the National Thyroid Research Project for Chinese Young and Middle-aged Doctors (2020), the Natural Science Foundation of Jiangsu Province (BK20200145).
Author information
Authors and Affiliations
Contributions
XX, HZ and JL conceived and designed the study. FW and JL supervised the study. LS, RD, XX and QJ performed the experiments and prepared the figures. LS and RD performed the statistical analysis. HZ, LS, ZS and WW contributed to the acquisition, analysis, or interpretation of the data. LS, ZS and RD contributed materials. XX, LS and RD drafted the paper. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The studies involving human participants were reviewed and approved by the Institutional Ethics Committees of the Affiliated Hospital of Jiangsu University, School of Medicine, Jiangsu University and Nanjing First Hospital, Nanjing Medical University. All participants provided informed consent, and their privacy was fully protected. All animal experiments were performed in accordance with the guidelines provided by the Animal Ethics Committee of Nanjing First Hospital (DWSY-22105231).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, L., Duan, R., Sun, Z. et al. LncRNA GLTC targets LDHA for succinylation and enzymatic activity to promote progression and radioiodine resistance in papillary thyroid cancer. Cell Death Differ 30, 1517–1532 (2023). https://doi.org/10.1038/s41418-023-01157-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01157-6
- Springer Nature Limited
This article is cited by
-
The roles and molecular mechanisms of non-coding RNA in cancer metabolic reprogramming
Cancer Cell International (2024)
-
Advances in metabolic reprogramming of NK cells in the tumor microenvironment on the impact of NK therapy
Journal of Translational Medicine (2024)
-
A novel tyrosine tRNA-derived fragment, tRFTyr, induces oncogenesis and lactate accumulation in LSCC by interacting with LDHA
Cellular & Molecular Biology Letters (2023)