Abstract
Exposure to artificial light at night (LAN) can induce obesity, depressive disorder and osteoporosis, but the pernicious effects of excessive LAN exposure on tissue structure are poorly understood. Here, we demonstrated that artificial LAN can impair developmental growth plate cartilage extracellular matrix (ECM) formation and cause endoplasmic reticulum (ER) dilation, which in turn compromises bone formation. Excessive LAN exposure induces downregulation of the core circadian clock protein BMAL1, which leads to collagen accumulation in the ER. Further investigations suggest that BMAL1 is the direct transcriptional activator of prolyl 4-hydroxylase subunit alpha 1 (P4ha1) in chondrocytes, which orchestrates collagen prolyl hydroxylation and secretion. BMAL1 downregulation induced by LAN markedly inhibits proline hydroxylation and transport of collagen from ER to golgi, thereby inducing ER stress in chondrocytes. Restoration of BMAL1/P4HA1 signaling can effectively rescue the dysregulation of cartilage formation within the developmental growth plate induced by artificial LAN exposure. In summary, our investigations suggested that LAN is a significant risk factor in bone growth and development, and a proposed novel strategy targeting enhancement of BMAL1-mediated collagen hydroxylation could be a potential therapeutic approach to facilitate bone growth.
Similar content being viewed by others
Introduction
Exposure to artificial light at night (LAN) is an increasing global health problem that afflicts over 80% of the world’s population [1]. Epidemiological evidence have shown that shift-workers exposed to LAN have high risks of metabolic abnormalities, cardiovascular diseases and depressive disorder, etc. [2,3,4]. Recent research demonstrated that even one night of light exposure can impair the level of blood glucose and increased insulin resistance in human [5]. Being a circadian disruption factor, LAN has been proven to alter the expression profiles of hormones, including melatonin and corticosterone, which seriously disrupts daily metabolism and circadian rhythms [6,7,8,9]. Even though accumulated scientific evidence have reported on the various pernicious effects of artificial LAN on endocrine function and physiological condition etc., whether or not LAN can induce substantial organic changes are still elusive. It has also been noticed that exposure to artificial LAN may impair bone metabolism and contribute to bone mass loss [10]. Hence, it is necessary to investigate the effects and underlying mechanisms of excessive LAN on the regulation of osteogenesis.
In mammals, optical signaling is critical for circadian rhythm synchronization and orchestration of diurnal physiological activities [11]. The suprachiasmatic nucleus (SCN) receives optical signals transmitted by intrinsically photosensitive retinal ganglion cells (ipRGC), which then generates the 24-h rhythmic secretion of hormones, including melatonin and adrenocorticotropic hormone (ACTH) that modulate specific circadian rhythms of peripheral tissues [12,13,14,15,16]. Circadian clock entrained by the light cycle has been proven to be a critical factor regulating the metabolism of growth plate cartilage [17, 18]. Our previous studies have demonstrated that metabolic activities associated with osteogenic centers exhibit circadian rhythms, manifested as fast DNA replication in the daytime due to active cell mitosis, alternating with matrix synthesis during the night [19]. Circadian rhythm disruption can markedly impair the correct sequence of chondrocyte differentiation and endochondral ossification [20]. However, the pathological processes and underlying molecular mechanisms by which LAN impairs cartilage extracellular matrix (ECM) formation within the developmental growth plate remains unelucidated.
Here, we showed that artificial LAN can suppress cartilage formation within the developmental growth plate by inhibiting collagen hydroxylation via the BMAL1/P4HA1 signaling pathway, which leads to collagen accumulation in the ER that in turn impedes bone formation. This study thus provides a new insight of LAN being a key limiting factor for height increment, which could in turn provide a novel strategy for promoting bone development via activation of the BMAL1/P4HA1 signaling axis.
Results
LAN disrupts growth plate cartilage ECM formation and causes ER dilation
We first established a LAN-exposure mouse model to assay the effects of artificial LAN on cartilage ECM formation in osteogenic centers (Fig. 1a). We found that LAN-exposure mice exhibit a shorter body length compared with normal LD12:12 mice (Fig. 1b). Micro-CT results showed reduced femur length of LAN-exposure mice, along with decreased bone volume per tissue volume (BV/TV), bone surface area per tissue volume (BS/TV), trabecular number (Tb.N), and increased inter-trabecular space (Tb.Sp) in the osteogenic centers of the femur (Fig. 1c, d and Extended Data Fig. 1b). The thickness of trabecular (Tb.Th) and cortical bones (Ct.Th) are not altered significantly by LAN (Extended Data Fig. 1a, b). Safranin O staining demonstrated that the proliferation zone length and proliferating chondrocyte number were diminished, accompanied by a decrease in the ratio of cartilage matrix in the proliferation zone from 80.73% to 67.91% (Fig. 1e–g). The expression of predominant matrix components type II collagen (COL2A1) and type VI collagen (COL6A1) decreased significantly, which was concomitant with a slight decrease in aggrecan core protein (ACAN) expression (Fig. 1h). We noticed that collagen without triple-helical configuration labeled by collagen hybridizing peptide 5-FAM conjugate (F-CHP) was much more in growth plate of LAN mice, indicating there are more unstable collagen (Extended Data Fig. 1c). Transmission electron microscope (TEM) imaging showed that LAN exposure reduced the amount of collagen fibers in ECM and increased the diameters of endoplasmic reticulum (ER) in chondrocytes (Fig. 1i, j). These findings thus indicate an atrophic phenotype of growth plate cartilage in the artificial LAN model.
a Schematic illustration of the experimental protocol. b Representative images of LD12:12 and LAN-exposure mice. c Representative Micro-CT images of distal femurs from 7-week-old LD12:12 and LAN-exposure mice. d Quantitative skeletal parameters of femurs from LD12:12 and LAN-exposure mice, including bone length, bone volume per tissue volume (BV/TV) and bone surface per tissue volume (BS/TV), showing the variation of endochondral ossification. (n = 5 per group). *P < 0.05, **P < 0.01. e–g Representative Safranin O staining shows the morphology of distal femur growth plate cartilage and quantitative analysis of proliferative zone length, cell number and ECM proportion in LD12:12 and LAN mice. *P < 0.05, **P < 0.01. Scale bars, 100 μm. The black area indicates the chondrocytes and the blue area indicates the cartilage ECM. h Immunofluorescent staining of COL2A1, COL6A1 and ACAN in femur growth plates from LD12:12 and LAN mice, respectively. Scale bars, 20 μm. i, j Representative TEM images of growth plate cartilage ECM and rough ER within chondrocytes and quantification of ER diameter from LD12:12 and LAN-exposure mice. ****P < 0.0001. Black arrows indicate the rough ER. In LAN-exposure mice, the rough ER was dilated abnormally. Scale bars, 500 nm.
Bmal1 knockout suppresses cartilage ECM formation and elicits ER dilation
It must be noted that LAN exposure is closely correlated with circadian rhythm disruptions in multiple-organs [21]. We then analyzed changes in the expression of classical clock genes within growth plate cartilage tissues from LAN mice. The qPCR data showed that LAN significantly reduced the mRNA and protein level of the aryl hydrocarbon receptor nuclear translocator-like protein 1 (Bmal1), accompanied by decreased mRNA levels of the circadian locomotor output cycles protein kaput (Clock) gene and increased mRNA levels of the period circadian protein homolog 2 (Per2) gene, as well as nuclear receptor subfamily 1 group D member 1 (Nr1d1) gene (Fig. 2a, b). Our previous studies have shown that BMAL1 plays critical roles in chondrocyte catabolic and anabolic metabolisms [22]. Taken together, these evidence suggested that BMAL1 may be the gatekeeper of chondrocyte homeostasis while disrupted by artificial LAN exposure.
a Relative mRNA expression of Bmal1, Clock, Per1 and Cry2 in LD12:12 and LAN mice (n = 3 per group). *P < 0.05, ***P < 0.001, ****P < 0.0001, ns, not significant. b Immunohistochemistry staining of BMAL1 in growth plate cartilage of distal femur. Scale bars, 100 μm. c–e Representative Safranin O staining images of distal femur growth plate cartilage and quantitative analysis of proliferative zone lengths, proliferative chondrocyte numbers and ECM proportions in WT, Bmal1+/− and Bmal1−/− mice (n = 5 per group). Scale bars, 100 μm. f–h Representative Safranin O staining images of femur growth plate cartilage and quantitative analysis of proliferative zone lengths, proliferative chondrocyte numbers and ECM proportions in Bmal1fl/fl and Twist2-Cre Bmal1fl/fl mice. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. i Immunofluorescence images of COL2A1 and KDEL within the proliferative zone of growth plate cartilage from WT and Bmal1−/− mice. j Immunofluorescence images of COL2A1 and KDEL within the proliferative zone in the femur of Bmal1fl/fl and Twist2-Cre Bmal1fl/fl mice. Scale bars, 20 μm. k Colocalization analysis of COL2A1 and KDEL by pearson’s correlation in WT and Bmal1−/− mice. **P < 0.01. l Colocalization analysis of COL2A1 and KDEL by pearson’s correlation in Bmal1fl/fl and Twist2-Cre Bmal1fl/fl mice. m Representative TEM images of growth plate ECM of wild type and Bmal1−/− mice. Scale bars, 500 nm. n Representative TEM images of rough ER within proliferative chondrocytes from wild type and Bmal1−/− mice. Black arrows indicate the rough ER. In Bmal1−/− mice, the rough ER was dilated abnormally. Scale bars, 500 nm. o Quantitative analysis of ER diameter from wild type and Bmal1−/− mice. ****P < 0.0001. p Representative TEM images of growth plate ECM of Bmal1fl/fl and Twist2-Cre Bmal1fl/fl mice. Scale bars, 500 nm. q Representative TEM images of rough ER within proliferative chondrocytes from Bmal1fl/fl Bmal1fl/fl Twist2-Cre mice. Black arrows indicate the rough ER. In Bmal1fl/fl Twist2-Cre mice, the rough ER was dilated abnormally. Scale bars, 500 nm. r Quantitative analysis of ER diameters from Bmal1fl/fl and Twist2-Cre Bmal1fl/fl mice. ****P < 0.0001.
To explore the role of BMAL1 in cartilage ECM construction, we successfully constructed a BMAL1-deficiency (Bmal1−/−) mouse model (Extended Data Fig. 2a). Notably, Bmal1+/− and Bmal1−/− mice exhibit shorter femur than their wild-type littermates (Extended Data Fig. 2b). The length of proliferative zone within the growth plate cartilage are also reduced accompanied by decreased cell number in proliferative zone in Bmal1+/− and Bmal1−/− mice (Fig. 2c, d). In particular, compared with the control samples, the proportion of ECM in the proliferative zone of Bmal1+/− and Bmal1−/− mice was reduced by about 10% and 20%, respectively. This evidence suggested that BMAL1 contributes to cartilage ECM construction (Fig. 2e). To rule out the systemic influence of whole-animal Bmal1 knockout, we also constructed a conditional Bmal1 knockout model in murine chondrocyte and their precursors (Bmal1fl/fl; Twist2-Cre). Alcian blue and alizarin red staining of E18.5 stage mouse embryos revealed significantly reduced ECM synthesis and delayed ossification of upper limb and rib cartilage in Bmal1fl/fl; Twist2-Cre mice (Extended Data Fig. 2c). A significant reduction was observed in the growth plate proliferative zone length and the ECM proportion of Bmal1fl/fl; Twist2-Cre mice (Fig. 2f–h). Immunofluorescent staining showed that both whole-animal knockout and conditional knockout of Bmal1 significantly induced the intracellular accumulation of COL2A1 and COL6A1, except for ACAN in chondrocytes (Fig. 2i–l and Extended Data Fig. 2d–g). TEM image analysis further suggested that loss of Bmal1 generates sparse extracellular collagen fiber distribution and contributes to ER dilation (Fig. 2m–r). These results thus indicate that BMAL1 deficiency in chondrocytes severely inhibits growth plate cartilage ECM formation.
BMAL1 deficiency inhibits collagen secretion and contributes to ER stress
Next, to explore how BMAL1 regulates cartilage ECM formation, we selectively knocked down BMAL1 expression (Fig. 3a) and examined the COL2A1 content of primary chondrocytes. We found that BMAL1 knockdown slightly reduced COL2A1 expression, while COL2A1 in the supernatants decreased significantly in chondrocytes (Extended Data Fig. 3a, b). Immunofluorescent staining further revealed that COL2A1 was mainly located inside the ER in Bmal1-knockdown chondrocytes (Fig. 3b), indicating insufficient collagen transport from ER to Golgi.
a Validation of Bmal1 knockdown using anti-BMAL1 antibody. (Green: BMAL1; blue: DAPI; Scale bar, 20 μm). b Immunofluorescence analysis of COL2A1 and KDEL in scramble and Bmal1-KD chondrocytes, respectively. (Green: COL2A1; red: KDEL; blue: DAPI; Scale bar, 20 μm). c Time-series immunofluorescence analysis of COL2A1 and GM130 in synchronized scramble and Bmal1-KD chondrocytes, respectively. (Green: COL2A1; red: GM130; blue: DAPI; Scale bar: 20 μm). Bottom: 3D surface plots of the immunofluorescent images. d Representative TEM images of ER and quantitative analysis of ER diameters in scramble and Bmal1-KD chondrocytes. Black arrows indicate the rough ER. Right: Quantitative analysis of mean ER diameters. ****P < 0.0001. Scale bar, 500 nm. e Protein levels of ER stress markers, including BiP and CHOP in scramble and Bmal1-KD chondrocytes. f Representative TEM images of ER and quantitative analysis of ER diameters in Bmal1-KD and Bmal1, Col2a1 double-knockdown chondrocytes. Black arrows indicate the rough ER. Right: Quantitative analysis of mean ER diameters. ****P < 0.0001. Scale bar, 500 nm. g Protein levels of BiP and CHOP in chondrocytes with Bmal1 and Col2a1 double-knockdown. *P < 0.05, **P < 0.01, ****P < 0.0001. h Western blot showed the CHOP expression in WT, Bmal1+/− and Bmal1−/− mice. i Representative immunofluorescence images of DHE and Caspase-3 in WT and Bmal1−/− mice. Scale bar, 20 μm.
To confirm whether BMAL1 deficiency induces defective transport of COL2A1 from ER to Golgi, we analyzed the temporal secretory process of COL2A1. In scramble synchronized chondrocytes, COL2A1 exhibited a significant overlap with the Golgi marker GM130 at 12–16 h after synchronization, upon secretion into the extracellular space (Fig. 3c). In contrast, BMAL1 downregulation prevented COL2A1 from reaching the Golgi apparatus, leading to excessive COL2A1 accumulation in the ER (Fig. 3c and Extended Data Fig. 3c). We also noticed that F-CHP staining was extensive in Bmal1-knockdown chondrocytes (Extended Data Fig. 3d). Meanwhile, the levels of ER stress markers endoplasmic reticulum chaperone BiP and DNA damage-inducible transcript 3 protein (CHOP) were markedly elevated, accompanied by ER dilation (Fig. 3d, e and Extended Data Fig. 3e). To further confirm whether ER stress in Bmal1-knockdown chondrocytes were caused by COL2A1, we knocked down COL2A1 expression. Simultaneous knockdown of BMAL1 and COL2A1 decreased the expression of ER stress markers in chondrocytes (Fig. 3f, g), indicating that ER stress is mainly caused by improperly folded COL2A1 within the ER. Immunofluorescent staining showed increased CHOP in growth plate of Bmal1+/− and Bmal1−/− mice, thus validating in vivo ER stress induction (Fig. 3h). Moreover, Bmal1 knockout led to a slight increase in oxidative stress and cell apoptosis (Fig. 3i). Concurrent with ER stress, BMAL1 downregulation increase the Beclin-1 (BECN1) and microtubule-associated protein 1 light chain 3 beta (MAP1LC3B) expression, thus activating ER-Phagy (Extended Data Fig. 3f). We used the autophagy inhibitor bafilomycin A1 to inhibit ER-Phagy and the results showed an increasing COL2A1 content in ER, indicating that ER-Phagy is responsible for degradation of unfolded COL2A1 (Extended Data Fig. 3g, h). The flow cytometry analysis result showed that inhibition of autophagy/ER-phagy in Bmal1-deficient cells resulted in higher cell apoptosis while the simultaneous knockdown of Bmal1 and Col2a1 could reduce cell apoptosis (Extended Data Fig. 3i), suggesting that inhibition of ER-phagy in Bmal1-deficient cells increased apoptosis due to excessive stress. These data thus suggested that BMAL1 deficiency blocks COL2A1 transport from ER to the Golgi apparatus and triggers subsequent ER stress and collagen degradation.
Impaired COL2A1 hydroxylation in BMAL1 deficient chondrocytes triggers ER stress in a P4HA1-dependent manner
To dissect the mechanisms by which BMAL1 downregulation leads to COL2A1 misfolding, we performed genome-wide RNA sequencing to characterize the transcriptional profile of BMAL1-knockdown chondrocytes. A total of 1699 differentially expressed genes (DEGs) were found in BMAL1-knockdown chondrocytes, including 816 upregulated genes and 813 downregulated genes (Fig. 4a). Gene ontology (GO) analysis revealed that biological pathways associated with ER stress, extracellular matrix organization, collagen fibril organization and circadian rhythm were enriched, in which collagen fibril organization was identified as the most significantly enriched process (Fig. 4b). As previously mentioned, BMAL1 may be involved in collagen fibril organization, thus controlling collagen secretion.
a Volcano plot showed the DEGs between scramble and Bmal1-KD chondrocytes (P < 0.01). b Gene ontology (GO) enrichment analysis of those DEGs in sequence data of scramble and Bmal1-KD chondrocytes. c Heatmap showing the DEGs of collagen fibril organization. d Relative mRNA analysis of P4ha1, Serpinh1 by qRT-PCR in scramble and Bmal1-KD chondrocytes. ***P < 0.001, ****P < 0.0001. e, f Protein expression levels of P4HA1 in WT, Bmal1+/− and Bmal1−/− mice. *P < 0.05. g Quantitative analysis of COL2A1 content in supernatants of Bmal1-KD chondrocytes with P4HA1 or SERPINH1 upregulation. **P < 0.01, ****P < 0.0001. h Hydroxyproline content analysis of WT and Bmal1−/− mice (n = 3 per group). i Hydroxyproline content analysis of LD12:12 and LAN mice (n = 3 per group). j Immunofluorescence analysis of COL2A1 and KDEL expression in Bmal1-KD chondrocytes with or without P4ha1 overexpression. Scale bar, 20 μm. Right: 3D surface plots of the immunofluorescent images. k Hydroxyproline content analysis of scramble and Bmal1-KD chondrocytes with or without P4ha1 overexpression. l Protein expression levels of BiP and CHOP in Bmal1-KD chondrocytes with or without P4ha1 overexpression. m Representative TEM images of rough ER and quantitative analysis of ER diameters in Bmal1-KD chondrocytes with P4ha1 overexpression. Scale bar, 500 nm.
Then, we focused on the DEGs of collagen fibril organization process, in particular that related to the transport process of COL2A1 from ER to Golgi, including prolyl 4-hydroxylase subunit alpha-1 (P4ha1), and serpin H1 (Serpinh1) (Fig. 4c and Extended Data Fig. 4a, b). qRT-PCR assays further verified that both P4ha1 and Serpinh1 decreased significantly (Fig. 4d). Then, we examined the expression of P4HA1 in the growth plate cartilages and the results showed that the P4HA1 expression are obviously decreased in Bmal1+/− and Bmal1−/− mice in comparison with WT mice (Fig. 4e, f). Overexpression of P4HA1, a major proline hydroxylase which is necessary for collagen proper folding and secretion [23], effectively increased the COL2A1 content in Bmal1-knockdown chondrocyte supernatants, while SERPINH1 upregulation failed to reverse this phenotype (Fig. 4g, Extended Data Fig. 4e). This suggests that P4HA1 is the major downstream molecule of BMAL1 regulating COL2A1 secretion. We also found that P4HA1 and hydroxyproline (pro-OH) were significantly reduced in the growth plate cartilage of the LAN-exposure model or Bmal1−/− mice (Fig. 4h, i, Extended Data Fig. 4c, d). P4HA1 downregulation blocks COL2A1 transportation form ER to Golgi and induces increased ER stress (Extended Data Fig. 4f–i). In addition, P4HA1 overexpression can promote proline hydroxyl modification and reduce COL2A1 retention and improper folding of collagen, accompanied by alleviated ER stress in Bmal1 knockdown chondrocytes (Fig. 4j–m and Extended Data Fig. 4j). Altogether, these results demonstrated that BMAL1-deficiency blocks the process of collagen secretion via downregulation of P4HA1, which is required for collagen proline hydroxyl modification.
Transcriptional regulation by BMAL1 induces rhythmic expression of P4HA1 in chondrocytes
As a typical canonical transcription factor [24], BMAL1 fulfills its function by activating the transcription of downstream genes. To gain more insight into the mechanisms by which BMAL1 regulated P4HA1, the JSPAR database was used to predict the potential transcriptional binding site for BMAL1 in the promoter of P4ha1 (Fig. 5a). We performed chromatin immunoprecipitation (ChIP) assay and the results showed that BMAL1 binds to the P4ha1 promoter (−538 to −412,127 bp) containing an E-box element (CAGGTG) (Fig. 5b), suggesting that P4ha1 transcription can be controlled by BMAL1. Furthermore, BMAL1 upregulation by SR1078 significantly promoted BMAL1 binding to the P4ha1 promoter and there was a dose-dependent increase in BMAL1 and P4HA1 expression was observed in vitro (Fig. 5c, d). Then we used dual-luciferase assay to evaluate the transcription competency of P4ha1 promoter in chondrocytes. The results demonstrated that the transcriptional activity of the P4ha1 promoter was inhibited to some extent in the mutants with E-box elements (Fig. 5e). Then, we observed that the levels of BMAL1, P4HA1 and pro-OH exhibited circadian oscillation and the peaks of BMAL1, P4HA1 and pro-OH were near ZT6, ZT12 and ZT12 in the growth plate cartilages, respectively (Fig. 5f, g). Consistent with in vivo results, Bmal1 and P4ha1 also exhibited circadian oscillation and the mRNA peaks of Bmal1 was near CT4-8 and P4ha1 was near CT8-12 (Fig. 5h). And P4HA1 protein, with a maximum near CT12, was later than the peak of BMAL1 which peaked at CT8 while BMAL1 knockdown decreased the expression levels of BMAL1 and P4HA1 (Fig. 5i). In addition, the hydroxyproline content is rhythmically expressed, while such rhythmic expression disappeared upon BMAL1 downregulation (Fig. 5j), thus indicating that circadian fluctuation of P4HA1 induced by BMAL1 maintains the physiological activities of collagen proline hydroxyl hydroxylation. Next, we upregulated P4HA1 in BMAL1-knockdown chondrocytes and found that there was increased COL2A1 transport towards the Golgi apparatus (Fig. 5k). Together, our data confirmed that BMAL1 mediated P4ha1 transcription drives rhythmic hydroxylation of collagen in chondrocytes.
a Schematic representation of P4ha1 promotor and predicted BMAL1 binding site. b ChIP-qPCR analysis of the interaction between BMAL1 and P4ha1 promotor. c ChIP-qPCR analysis of the interaction between BMAL1 and P4ha1 promotor after SR1078 administration. ***P < 0.001. d Protein fold change analysis of BMAL1 and P4HA1 expression in chondrocytes treated with SR1078. e Dual-luciferase analysis of BMAL1 promoting P4ha1 transcription. f Protein levels of BMAL1 and P4HA1 within the growth plates of 7-week-old LD12:12 mice. g Hydroxyproline content analysis of the growth plates in 7-week-old LD12:12 mice over a 24-h period. h qRT-PCR analysis of the relative expression of P4ha1 and Bmal1 over a 24-h period in primary chondrocytes after synchronization. i Protein levels of BMAL1 and P4HA1 in scramble and Bmal1 deficient chondrocytes over a 24-h period. j Hydroxyproline content analysis of scramble and Bmal1-KD chondrocytes over a 24-h period after synchronization. k Time-series immunofluorescence analysis of COL2A1 and GM130 in synchronized Bmal1-KD mice with P4ha1 upregulated chondrocytes. (Green: COL2A1; red: GM130; blue: DAPI; Scale bar: 20 μm). Down direction: 3D surface plots of the immunofluorescent images.
Restoration of the BMAL1/P4HA1 axis reverses LAN-induced dysregulation of growth plate cartilage formation
To further validate whether decreased BMAL1/P4HA1 signaling axis is responsible for the dysregulation of cartilage ECM formation observed in the LAN-exposure mouse model, we performed in vivo experiments involving supplementation of SR1078, a small molecule which can promote BMAL1 expression (i.p. daily, 15 mg/kg) (Fig. 6a). To test whether SR1078 affects the BMAL1/P4HA1 axis, we detected the mRNA expression levels of Bmal1 and P4ha1 over a 24-h period and found that the mRNA expression levels of Bmal1 and P4ha1 follow the circadian rhythm, with P4ha1 reaching its trough around ZT16 in the femur growth plate of LD12:12 mice, while its oscillation disappeared in the LAN-exposure model. SR1078 treatment effectively recovered the circadian rhythm and expression of Bmal1 and P4ha1, as well as hydroxyproline content in the distal femur growth plate (Fig. 6b–d). And our result showed that increased Bmal1 level in growth plate cartilages was detected as early as 6 h post injection and peaked at 12-h post injection and the Bmal1-inducing effect by SR1078 can last at least 24 h (Extended Data Fig. 5a). Then we tested the bone formation activity in vivo and found that SR1078 significantly elevated bone length, BV/TV, BS/TV and Tb.N in LAN model (Fig. 6e and Extended Data Fig. 5b). This indicates that restoration of the BMAL1/P4HA1 axis can rescue LAN exposure-induced defects in bone formation. LAN mice displayed a reduction in body weight after 12 days of exposing to LAN and body weight increased after SR1078 treatment, while the food consumption did not differ among all groups (Extended Data Fig. 5c, d), indicating that change of bone volume is not caused by food intake. Moreover, we found that SR1078 treatment increased cartilage matrix mass in the proliferative zone, effectively enhancing the ECM ratio (Fig. 6f), accompanied by elevated extracellular collagen content, increased amount of collagen fibers and reversal of ER dilation (Fig. 6g, h and Extended Data Fig. 5e). Collectively, these results demonstrated that upregulation of BMAL1/P4HA1 effectively promotes cartilage ECM deposition and subsequent endochondral ossification after LAN exposure.
a Schematic illustration of the experimental protocol. b, c qRT-PCR analysis of the relative expression of P4ha1 and Bmal1 over a 24-h period within the growth plates of 7-week-old LD12:12 and LAN-exposure mice, with or without SR1078 administration (n = 5 per group). d Hydroxyproline content analysis of growth plate cartilage in LD12:12, LAN and LAN-exposure mice treated with SR1078. *P < 0.05, **P < 0.01. e Micro-CT and quantitative skeletal parameters of femur in LD12:12 and LAN-exposure mice with or without SR1078 administration. f Safranin O staining of femur growth plate cartilage and quantitative analysis of proliferative zone length and ECM proportion in LD12:12 and LAN-exposure mice with or without SR1078 administration. *P < 0.05. g Immunofluorescence analysis of COL2A1 AND KDEL in the femur of LD12:12 and LAN-exposure mice, with or without SR1078 administration. Scale bar: 20 μm. h Representative TEM images of cartilage ECM and rough ER within proliferative chondrocytes of LD12:12 and LAN-exposure mice, with or without SR1078 administration. Black arrows indicate the rough ER.
Discussion
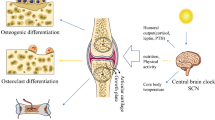
Our study demonstrates that decreased BMAL1/P4HA1 signaling drives the deficiency of collagen hydroxylation which accounts for the impairment of cartilage ECM formation induced by artificial LAN exposure (Fig. 7). Previous report indicated that short term LAN also decreases ovarian follicle number, accompanied with downregulated clock gene expression and DNA damage [25]. Circadian clock is entrained by light signals and LAN impairs melatonin level, sleep quality and circadian rhythm [6, 26, 27]. Upon receiving light signals from retina ipRGCs, SCN synchronizes the peripheral tissue clock, while untimely light stimuli can disrupt local circadian clock gene expression [28]. Therefore, LAN-associated health risks are worthy of more attention and everyone should be cautioned to avoid exposure to artificial LAN, especially adolescents.
BMAL1 downregulation induced by LAN markedly inhibits P4ha1 transcription in chondrocytes. Less P4HA1 caused insufficient collagen prolyl hydroxylation, leading to ER stress and inhibition of COL2A1 secretion. Thus, cartilage ECM formation within the growth plate was suppressed by artificial LAN exposure (Created with BioRender.com).
The circadian clock is an essential regulator of growth plate cartilage homeostasis and cartilage development [17, 18]. Our recent study demonstrated that the cartilage circadian clock is predominantly regulated by melatonin-mediated AMP-activated protein kinase (AMPK) signaling pathway [19]. Among the common circadian clock genes, we found that Bmal1 was significantly downregulated by LAN. BMAL1 has been proven to be responsible for maintaining cartilage circadian rhythms and BMAL1-deficiency in chondrocytes could induce progressive degeneration characterized by loss of cartilage [22]. We have also examined the morphology of the growth plate from the 6-month-old Twist2-specific Bmal1-KO mice. We didn’t observe the “cavity-like” structural changes in the growth plate (Extended Data Fig. 2h). Unlike growth plate, articular cartilage is a load-bearing tissue, the mechanical loading likely account for this discrepancy. Previous report indicated that Bmal1 deletion inhibits cell proliferation and increases cell apoptosis in the growth plate cartilages, which affects growth plate thickness [29]. Downregulation of BMAL1 aggravates catabolism and local inflammation in the pathophysiology of cartilage degeneration related disorders [30, 31]. In this study, we chose Twist2-specific Bmal1-knockout mice, in which the Twist2-cre mice have been conventionally used to achieve gene inactivation in cartilages [20, 32], to investigate the roles of BMAL1 in the whole process of cartilage differentiation and development. We noticed that Bmal1-deficiency in chondrocytes could decrease the thickness of the proliferative zone, which is closely associated with reduced COL2A1 secretion and impaired cartilage matrix formation. Moreover, the previous study reported that postnatal deletion of Bmal1 in matured osteoblasts can delay endochondral ossification, resulting in a thicker tibial growth plate [33]. In the bone marrow, osteoblast-specific Bmal1 knockout can increase trabecular bone mass through activation of BMP2/SMAD1 signaling pathway [33]. Overall, BMAL1 in chondrocytes is vital for chondrocyte sequence differentiation and matrix formation, and osteoblast BMAL1 also plays an important indirect regulatory role in endochondral ossification, suggesting that BMAL1 regulates physiological activities of growth plate cartilage through multiple mechanisms.
Here, we revealed that BMAL1 deficiency induces defects in collagen assembly through P4HA1. P4HA1 is responsible for 4-hydroxyproline generation in the Yaa position of Gly-Xaa-Yaa repeats of collagen, which is necessary for collagen triple-helix structure formation [23, 34, 35]. Individuals with bi-allelic P4HA1 mutations display severe bone dysplasia and muscle weakness, and loss of P4HA1 is known to induce embryonic lethality in mice [35, 36]. We further confirmed that BMAL1 modulates P4ha1 expression through direct transcriptional regulation, which leads to rhythmic fluctuations in P4HA1 and hydroxyproline expression, which peaks around CT12. This result correlates exactly with our recent findings that cartilage matrix synthesis mainly occurs during night time [19]. As reported by a previous study, the circadian clock precisely regulates collagen secretion in tendon via traffic-related genes, including Sec12a, Mia3, Pde4d and Vps33b [37]. We observed that COL2A1 reach the Golgi by CT12 to CT16 in chondrocytes, which is coincident with the timing of collagen hydroxylation, thus indicating that COL2A1 hydroxylation and transportation are intimately coupled together. Therefore, prolyl hydroxylases regulate collagen hydroxylation and secretion in a circadian-dependent manner and LAN disrupted BMAL1-driven collagen hydroxylation leads to impaired matrix synthesis and cartilage formation at night.
We have showed that upregulation of BMAL1-driven collagen hydroxylation by the small molecule SR1078 effectively alleviated the detrimental impacts of LAN exposure in mice. Emerging evidences suggest that both hormones (including melatonin, thyroxine, somatotropin, etc.) secretion and collagen synthesis are orchestrated by intrinsic circadian clock system [38]. The ubiquitous LAN can disrupt the intrinsic circadian rhythms and then destroy the secretion of melatonin, glucocorticoid hormones, gonadotropins and thyroid hormones [39]. These findings suggested that circadian rhythm targeting agonist SR1078 could improve most growth disorders through restoring the daily synthesis and secretion of hormones and collagen. In addition, BMAL1 downregulation has been proven in human OA cartilage and the articular cartilage of aged mouse [22, 40]. Bmal1 ablation in mouse chondrocytes could abolish chondrocyte intrinsic circadian rhythm and cause OA-like degeneration [22]. Conversely, BMAL1 overexpression neutralize the IL-1β-induced degeneration of chondrocyte and promote COL2A1 and ACAN expression and inhibit MMP13 and ADAMTS5 expression [41, 42]. Therefore, we believe that both impaired growth and OA could be treated by SR1078. The Bmal1-inducing effect by SR1078 in first 24 h post injection showed 2- to 3-fold than that in second 24 h post injection, suggesting that once per day dosing is preferred. To compliant with the intrinsic BMAL1 expression pattern, the time point of SR1078 application should be ZT18 in mice. Moreover, we recommend that SR1078 can be applied until the end of growth plate development. In the future, the efficacy of SR1078 treatment of LAN exposure needs to be evaluated in clinical trials, and the medication safety needs to be rigorously evaluated along with its efficacy.
Materials and methods
Animal study
Three-week-old male C57BL/6J mice obtained from Vital River Bioscience (Beijing, China) were used in this study. Mice were randomly assigned to either the LD12:12 or LAN-exposure group after acclimating to the environment for one week. LD12:12 mice were maintained in a 12 h light/12 h dark (LD 12:12) cycle. Whereas extra light was administered from zeitgeber time (ZT)13 to ZT19 in the LAN-exposure group. The mice were then sacrificed for femur extraction 3 weeks later. The Bmal1−/− and Bmal1fl/fl Twist2-Cre mouse have been described previously [20]. The Bmal1+/− mice were bred for production of Bmal1−/− mice. The identified primer sequences were: 5′-CCAC CAAGCCCAGCAACTCA-3′ (common forward), 5′-ATTCGGCCCCCTATCTTCTGC-3′ (wild-type reverse), 5′-TCGCCTTCTATCGCCTTCTTGACG-3′ (targeting reverse). We crossed Bmal1fl/fl mice with Twist2-Cre mice to generate mice lacking Bmal1 in chondrocytes. For SR1078 treatment, SR1078 (MCE, HY-14422) at a concentration of 15 mg/kg was administrated (i.p. daily) for three weeks. Experimenter blinding was done at the time of group allocation before and after animal model establishment. We excluded the mice with poor health in this study. The experimental protocol was approved by the Institutional Animal Care and Use Committee of Tongji Medical College (LAUCU Number:2809).
Cell culture and reagent
Primary chondrocytes were isolated from the articular cartilage of 3-week-old male SD rat. Briefly, the distal femurs were extracted and the soft tissues were cleaned up. After 3 times washing with PBS, the cartilage tissue was separated and cut into pieces as small as can be obtained. After 30 min digestion with 0.25% (w/v) trypsin at 37 °C, collagenase II solution was mixed with the samples at 37 °C overnight. The next day, after centrifugation, the precipitate was suspended in DMEM/F12 (Hyclone) medium with 10% (v/v) FBS (Gibco). Primary chondrocytes were cultured at 37 °C, within a humidified 5% CO2 atmosphere. For BMAL1 inhibition, chondrocytes were treated with SR9009 (MCE, HY-16989) at 10μmol/mL. For inhibition of autophagy, chondrocytes were treated with Bafilomycin A1 (HY-100558).
Micro-CT analysis
The mice femurs were extracted and fixed in 4% (w/v) paraformaldehyde. The samples were positioned at the scanning bed and scanned with a spatial resolution of 9 μm per voxel by Micro-CT (SkyScan 1176, Broker). We performed Micro-CT analysis and calculated the bone morphology parameters including bone length, bone volume/total volume, trabecular number, trabecular separation, trabecular thickness and bone surface area/bone volume by the InstaRecon/NRecon Research Workplace software.
Transcript sequencing analysis
Total RNA of primary chondrocytes with or without Bmal1 knockdown were isolated by RNAiso Plus (Invitrogen, USA). A minimum of 1 μg RNA were sent to Nonvogene (Wuhan, China) for quality control, library preparation and next-generation sequencing. Briefly, the RNA was fragmented into 250–300 bp fragment and reverse transcribed. The selected cDNA was subjected to PCR with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. Then, the cDNA was purified by the AMPure XP system. Transcriptome sequencing was performed with the Novaseq platform (Illumina, San Diego, CA, USA), which generated 150 bp paired-end reads. The read numbers were counted by featureCounts v1.5.0-p3, followed by calculation of FPKM of each gene. We then used the DESeq2 R package (1.16.1) to perform differential expression analysis of the two groups.
Histology and immunohistochemistry analyses
The mice femurs were fixed in paraformaldehyde, EDTA (10%)-decalcified and paraffin-embedded. Then the samples were deparaffinized and rehydrated in xylene and graded ethanol, respectively. The 6-μm paraffin sections were stained with fast green and safranin O following standard protocols. To measure the ECM proportion, we divided the area of chondrocytes and extracellular matrix in the images of Safranin O staining according to literature method [43, 44]. Then the software Image J was used to calculate the area of chondrocyte and extracellular matrix respectively in each section. The ECM/chondrocyte proportion in growth plate was defined as the ratio of ECM/cell area to total area. For immunohistochemistry, antigen retrieval was performed with citrate buffer. The BMAL1 levels in the growth plate were detected using BMAL1 antibody (1:200, Abcam, ab93806). After incubation with BMAL1 antibody overnight at 4 °C, the sections were incubated with biotin-conjugated secondary antibody, and then visualized using a DAB kit and hematoxylin counterstaining. Finally, the sections were sealed with neutral gum and the images were captured under a Nikon microscope (Japan).
Cell synchronization
Cell circadian rhythm establishment was conducted as described previously [45]. Briefly, the primary chondrocytes were cultured in cell culture dishes until the cell confluence reached 100%. Subsequently, the cells were treated with dexamethasone (1 μmol/L, Sigma) for 1 h and the medium was changed to complete DMEM/F12 medium, at which the moment was set as CT0. The cells were then harvested for further experiments every 6 h.
Western blotting
The relative protein expression levels were analyzed by western blotting. The cell samples were lysed with RIPA buffer containing protease inhibitor cocktail and phosphatase inhibitor. The mixture was then centrifuged at 12,000 g at 4 °C after cell lysis with a ultrasonicator. Then the protein concentrations were determined by a Bicinchoninic Acid Kit (Beyotime, P0012S). Meanwhile, the SDS loading buffer (5×) was then added into the supernatant for 5 min at 95 °C. A total of 30 μg protein within each lane were separated using 10% (w/v) polyacrylamide gels and then transferred to a PVDF membrane. The membranes were first blocked by 5% (w/v) skimmed milk, and the various proteins were detected by incubation with primary antibodies, including BMAL1 (1:1000, Abcam, ab93806), COL2A1 (1:1000, Abcam, ab34712), BiP (1:1000, Proteintech, 11587-1-AP), CHOP (1:1000, Proteintech, 15204-1-AP), P4HA1 (1:4000, Proteintech, 12658-1-AP), β-actin (1:1000, Proteintech, 20536-1-AP), GAPDH (1:10000, Proteintech, 10494-1-AP) overnight at 4 °C. Next, the membranes were visualized by secondary antibody and ECL solution. The antibodies used in this study are listed in Supplementary Table 1.
QRT-PCR
RNA was isolated from mice growth plate cartilage and cells as described previously [19]. Briefly, the tissues and cells were mixed in RNAiso Plus (Invitrogen, USA) and RNA was isolated according to the manufacturer’s protocol. Then the extracted RNA was reverse transcribed using HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China). The PCR amplification quantitative analyses were performed with ChamQTM SYBR® qPCR Master Mix (Vazyme, China). GAPDH expression was used for normalization and the primer sequences are listed in Supplementary Table 2.
Immunofluorescence staining and Collagen-hybridization peptide assays
For tissue immunofluorescence staining, antigen retrieval and blocking were performed on rehydrated cartilage sections. Cells were washed by PBS and fixed with 4% (w/v) paraformaldehyde for 20 min, followed by permeabilization with 0.1% (w/v) Triton-X solution. After 3 times washing in PBS, the samples were blocked with 5% (w/v) BSA solution for 1 h. COL2A1 was then detected using mouse mAb (1:100; Invitrogen, MA5-12789) and rabbit pAb (1:100, Abcam, ab34712), ACAN was detected using rabbit mAb (1:200; Proteintech, 13880-1-AP), COL6A1 was detected using rabbit pAb (1:200; Proteintech, 17023-1-AP), P4HA1 was detected using rabbit pAb (1:200; Proteintech, 12658-1-AP), Caspase3 was detected using rabbit pAb (1:200; Proteintech, 19677-1-AP), ER was detected using mouse mAb to KDEL (1:200; Abcam, ab12223) or rabbit pAb (1:200; Introvigen, PAI-013) and cis-Golgi was detected using rabbit pAb to GM130 (1:200; Introvigen, PA5-95727). Secondary antibodies conjugated to 488 and Cy3 (1:200; Proteintech) were used. Pearson’s correlation was measured by plugin Coloc2 in Image J used to performed the colocalization analysis of COL2A1/ACAN/COL6A1 and KDEL. For ROS detection, dihydroethidium (DHE) (Sigma) staining was performed on fresh growth plate sections.
For detection of unfolded collagen chains in vitro, Collagen Hybridizing Peptide, 5-FAM Conjugate (F-CHP, 3Helix) were used according to the manufacturer’s instructions. Briefly, the F-CHP solution was heated at 80 °C for 5 min, followed by exposure to an ice-water bath for 30 s. Then the cells were incubated with F-CHP at 4 °C overnight. The cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Images were captured under a Nikon laser scanning confocal microscope and NIS-Elements Viewer software.
Transmission electron microscopy
The thin slice of growth plate cartilage or primary chondrocytes were harvested and fixed in 0.1 mol/L sodium cacodylate buffer containing 2.5% (w/v) glutaraldehyde for 2 days at 4 °C. Then the samples were fixed again in 1% (w/v) osmium tetroxide for 1 h, and then dehydrated by graded ethanol. After paraffin embedment and preparation of ultrathin sections (60–80 nm), the samples were examined by a FEI Tecnai G2 20 TWIN transmission electron microscope (FEI Company).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays (ChIP) was performed using a chromatin immunoprecipitation kit (Beyotime), according to the standard protocol as described before. After cross-linking, lysis, disruption into DNA fragments, 10% was kept as input. Protein A/G agarose beads and BMAL1 antibody (Abcam, ab3350, 1 μg) were mixed with the supernatants overnight at 4 °C. Then, the Immunoprecipitated complexes were eluted for further PCR analysis. The primer sequences used for this experiment was 5′-CAGACCTAGATGCCGGGAAG-3′ (forward) and 5′-TTTCGCTTAGTCTCC CATCCG-3′ (reverse), yielding a 127 bp product (−538 to −412).
Viral infection, RNA interference and plasmid transfection
For viral infection, the CRISPR/CAS9 system (pLV[Exp]-CBh>hCas9, VB160923-1033trt, REBIOSCI SCIENTIFIC, China) with lentiviral vector (pLV[2gRNA]-Puro-U6, VB180124-1097pnr, REBIOSCI SCIENTIFIC, China) was used to knockdown Bmal1. Lentiviral vector (U6-MCS-Ubiquitin-Cherry-IRES-puromycin, GV298, GENECHEM Biosciences, China) containing Col2a1-RNAi (NM_012929) were used to knockdown Col2a1. We used a CRISPR/CAS9 system (YCas-LV002) and lentiviral vector (YKO-LV004-P4ha1[gRNA1-gRNA2]) to knockdown P4ha1. Lentiviral vector (Ubi-MCS-IRES-Hygromycin, CV237, GENECHEM Biosciences, China) containing rP4ha1 (NM_172062) gene sequence was used to overexpress P4ha1. For Serpinh1 overexpression, we used a lentiviral vector (Ubi-MCS-SV40-blasticidin, CV301, GENECHEM Biosciences, China) containing rSerpinh1 (NM_017173) gene sequence. Oligonucleotide sequences are detailed in Supplementary Table 4. The virus (multiplicity of infection of 10) was incubated with primary chondrocytes for 12 h, then the medium was replaced by fresh DMEM/F12 supplemented with 10% (v/v) FBS.
For plasmid transfection, firefly luciferase reporter vector and pRL-TK plasmids were constructed by TsingKe Biological Technology (Wuhan, China). The pGL3-Basic, pGL3-Basic P4ha1 plasmid, as well as site-mutant or site-deleted plasmid were transfected into the cells for further luciferase report assay. Briefly, the firefly luciferase reporter plasmid (375 ng) and pRL-TK plasmids (125 ng) were mixed with Lipo8000™ Transfection Reagent (Beyotime) in serum-free medium, then the mixtures were transfected into the cells.
Luciferase reporter assay
The pGL3-basic vector containing the P4ha1 promotor (2.0 kb) were constructed. Based on the pGL3-P4ha1 promoter (2.0 kb), we further constructed site-mutant plasmids in which the E-box like sequence CCAGGTGACA (-468 to -477 bp) were replaced by CTGGGCAACA and site-deleted plasmid in which the region was deleted. Primary chondrocytes were seed into a 24-well plate and cultured until 75% confluence were obtained. The firefly luciferase reporter plasmid (375 ng) and pRL-TK plasmids (125 ng) were co-transfected into the cells by Lipo8000™ Transfection Reagent (Beyotime, 0.8 μL/well). After 48 h incubation, the cells were lysed with the lysis buffer and the Fluc and Rluc activities were measured via the Dual Luciferase Reporter Assay System (Promega) according to standard protocols. The Fluc/Rluc ratio were calculated to assess the relative luciferase signal.
Elisa
For measurement of secreted collagen alpha-1(II) chain of chondrocytes, we replaced the medium with serum-free medium when the confluence of chondrocytes reached 100%. After 24 h of culture, the cell supernatants form primary chondrocytes were collected. The ELISA kit (CUSABIO, China) was used to quantify the COL2A1 secretion according to the instruction.
Prolyl hydroxylation assay
Collagen prolyl hydroxylation content was measured using the prolyl hydroxylation kit (Sigma-Aldrich, MAK008) according to the manufacturer’s protocol. Briefly, the samples were mixed with 100 μL 6 mol/L HCl. After hydrolysis at 120 °C for 3 h, the tubes were centrifuged at 10,000 × g for 3 min. The hydroxylation standards (0, 0.2, 0.4, 0.6, 0.8, and 1.0 μg) were then added into the 96-well plate. The samples were added into the 96-well plate at 40 μL per well, and the plate were evaporated to dryness. Then 100 μL Chloramine T/Oxidation buffer mixture were placed in each well and incubated for 5 min. Subsequently, 100 μL of diluted DMAB reagent were added and heated at 60 °C for 90 min, and then the absorbance at 560 nm were measured.
Statistics and reproducibility
No statistical methods were used to pre-determine sample size, but our sample sizes were similar to previous studies. All quantitative data are presented as mean ± standard deviation. All statistical analysis was performed using GraphPad Prism 7.0 software. The numbers of animals in each experiment are stated in the figure legends. All in vitro experiments were repeated independently at least 3 times. We used Shapiro–Wilk test to examine the normal distribution of the data. Statistical analysis of comparision between two groups was performed by the Student’s t-test. For multiple groups, one-way ANOVA with the Bonferroni correction was used for analysis of statistical significance. A P value < 0.05 was considered significant.
Data availability
All data supporting the results are available from the corresponding author upon request.
References
Falchi F, Cinzano P, Duriscoe D, Kyba CC, Elvidge CD, Baugh K, et al. The new world atlas of artificial night sky brightness. Sci Adv. 2016;2:e1600377.
Münzel T, Hahad O, Daiber A. The dark side of nocturnal light pollution. Outdoor light at night increases risk of coronary heart disease. Eur Heart J. 2021;42:831–4.
Navara KJ, Nelson RJ. The dark side of light at night: physiological, epidemiological, and ecological consequences. J Pineal Res. 2007;43:215–24.
An K, Zhao H, Miao Y, Xu Q, Li YF, Ma YQ, et al. A circadian rhythm-gated subcortical pathway for nighttime-light-induced depressive-like behaviors in mice. Nat Neurosci. 2020;23:869–80.
Mason IC, Grimaldi D, Reid KJ, Warlick CD, Malkani RG, Abbott SM, et al. Light exposure during sleep impairs cardiometabolic function. Proc Natl Acad Sci USA. 2022;119:e2113290119.
Fonken LK, Nelson RJ. The effects of light at night on circadian clocks and metabolism. Endocr Rev. 2014;35:648–70.
Tähkämö L, Partonen T, Pesonen AK. Systematic review of light exposure impact on human circadian rhythm. Chronobiol Int. 2019;36:151–70.
Haraguchi S, Kamata M, Tokita T, Tashiro KI, Sato M, Nozaki M, et al. Light-at-night exposure affects brain development through pineal allopregnanolone-dependent mechanisms. eLife. 2019;8:e45306.
Bedrosian TA, Galan A, Vaughn CA, Weil ZM, Nelson RJ. Light at night alters daily patterns of cortisol and clock proteins in female Siberian hamsters. J Neuroendocrinol. 2013;25:590–6.
Feskanich D, Hankinson SE, Schernhammer ES. Nightshift work and fracture risk: the Nurses’ Health Study. Osteoporos Int. 2009;20:537–42.
Panda S. Circadian physiology of metabolism. Science. 2016;354:1008–15.
Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49.
Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, Castrucci AM, et al. Melanopsin is required for non-image-forming photic responses in blind mice. Science. 2003;301:525–7.
Ishida A, Mutoh T, Ueyama T, Bando H, Masubuchi S, Nakahara D, et al. Light activates the adrenal gland: timing of gene expression and glucocorticoid release. Cell Metab. 2005;2:297–307.
Liu Z, Gan L, Luo D, Sun C. Melatonin promotes circadian rhythm-induced proliferation through Clock/histone deacetylase 3/c-Myc interaction in mouse adipose tissue. J Pineal Res. 2017;62:e12383.
So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci USA. 2009;106:17582–7.
Schilperoort M, Bravenboer N, Lim J, Mletzko K, Busse B, van Ruijven L, et al. Circadian disruption by shifting the light-dark cycle negatively affects bone health in mice. FASEB J. 2020;34:1052–64.
Yuan G, Hua B, Yang Y, Xu L, Cai T, Sun N, et al. The circadian gene clock regulates bone formation via PDIA3. J Bone Miner Res. 2017;32:861–71.
Yu S, Tang Q, Chen G, Lu X, Yin Y, Xie M, et al. Circadian rhythm modulates endochondral bone formation via MTR1/AMPKβ1/BMAL1 signaling axis. Cell Death Differ. 2022;29:874–87.
Yu S, Tang Q, Xie M, Zhou X, Long Y, Xie Y, et al. Circadian BMAL1 regulates mandibular condyle development by hedgehog pathway. Cell Prolif. 2020;53:e12727.
Tam SKE, Brown LA, Wilson TS, Tir S, Fisk AS, Pothecary CA, et al. Dim light in the evening causes coordinated realignment of circadian rhythms, sleep, and short-term memory. Proc Natl Acad Sci USA. 2021;118:e2101591118.
Dudek M, Gossan N, Yang N, Im HJ, Ruckshanthi JP, Yoshitane H, et al. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J Clin Investig. 2016;126:365–76.
Pihlajaniemi T, Myllylä R, Kivirikko KI. Prolyl 4-hydroxylase and its role in collagen synthesis. J Hepatol. 1991;13 Suppl 3:S2–7.
Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, et al. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science. 2012;337:189–94.
Li Y, Cheng S, Li L, Zhao Y, Shen W, Sun X. Light-exposure at night impairs mouse ovary development via cell apoptosis and DNA damage. Biosci Rep. 2019;39:BSR20181464.
Gooley JJ, Chamberlain K, Smith KA, Khalsa SB, Rajaratnam SM, Van Reen E, et al. Exposure to room light before bedtime suppresses melatonin onset and shortens melatonin duration in humans. J Clin Endocrinol Metab. 2011;96:E463–472.
deHaro D, Kines KJ, Sokolowski M, Dauchy RT, Streva VA, Hill SM, et al. Regulation of L1 expression and retrotransposition by melatonin and its receptor: implications for cancer risk associated with light exposure at night. Nucleic Acids Res. 2014;42:7694–707.
Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–5.
Ma Z, Jin X, Qian Z, Li F, Xu M, Zhang Y, et al. Deletion of clock gene Bmal1 impaired the chondrocyte function due to disruption of the HIF1α-VEGF signaling pathway. Cell Cycle. 2019;18:1473–89.
Chen G, Zhao H, Ma S, Chen L, Wu G, Zhu Y, et al. Circadian rhythm protein Bmal1 modulates cartilage gene expression in temporomandibular joint osteoarthritis via the MAPK/ERK pathway. Front Pharmacol. 2020;11:527744.
Peng P, Wang D, Xu X, Wang D, Gao B, Wang H, et al. Targeting clock-controlled gene Nrf2 ameliorates inflammation-induced intervertebral disc degeneration. Arthritis Res Ther. 2022;24:181.
Abou-Jaoude A, Courtes M, Badique L, Elhaj Mahmoud D, Abboud C, Mlih M, et al. ShcA promotes chondrocyte hypertrophic commitment and osteoarthritis in mice through RunX2 nuclear translocation and YAP1 inactivation. Osteoarthr Cartil. 2022;30:1365–75.
Qian Z, Zhang Y, Kang X, Li H, Zhang Y, Jin X, et al. Postnatal conditional deletion of Bmal1 in osteoblasts enhances trabecular bone formation via increased BMP2 Signals. J Bone Miner Res. 2020;35:1481–93.
Taga Y, Kusubata M, Ogawa-Goto K, Hattori S. Stable isotope-labeled collagen: a novel and versatile tool for quantitative collagen analyses using mass spectrometry. J Proteome Res. 2014;13:3671–8.
Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivirikko KI, et al. Loss of assembly of the main basement membrane collagen, type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J Biol Chem. 2007;282:2512–9.
Zou Y, Donkervoort S, Salo AM, Foley AR, Barnes AM, Hu Y, et al. P4HA1 mutations cause a unique congenital disorder of connective tissue involving tendon, bone, muscle and the eye. Hum Mol Genet. 2017;26:2207–17.
Chang J, Garva R, Pickard A, Yeung CC, Mallikarjun V, Swift J, et al. Circadian control of the secretory pathway maintains collagen homeostasis. Nat Cell Biol. 2020;22:74–86.
Gamble KL, Berry R, Frank SJ, Young ME. Circadian clock control of endocrine factors. Nat Rev Endocrinol. 2014;10:466–75.
Ouyang JQ, Davies S, Dominoni D. Hormonally mediated effects of artificial light at night on behavior and fitness: linking endocrine mechanisms with function. J Exp Biol. 2018;221:jeb156893.
Akagi R, Akatsu Y, Fisch KM, Alvarez-Garcia O, Teramura T, Muramatsu Y, et al. Dysregulated circadian rhythm pathway in human osteoarthritis: NR1D1 and BMAL1 suppression alters TGF-β signaling in chondrocytes. Osteoarthr Cartil. 2017;25:943–51.
Yang W, Kang X, Liu J, Li H, Ma Z, Jin X, et al. Clock gene Bmal1 modulates human cartilage gene expression by crosstalk with Sirt1. Endocrinology. 2016;157:3096–107.
He T, Pang S, Wang H, Yun H, Hao X, Jia L, et al. Drugging the circadian clock feedback cycle to ameliorate cartilage degeneration. FEBS J. 2022;289:6643–58.
Zhang M, Lu Q, Egan B, Zhong XB, Brandt K, Wang J. Epigenetically mediated spontaneous reduction of NFAT1 expression causes imbalanced metabolic activities of articular chondrocytes in aged mice. Osteoarthr Cartil. 2016;24:1274–83.
Stegen S, Laperre K, Eelen G, Rinaldi G, Fraisl P, Torrekens S, et al. HIF-1α metabolically controls collagen synthesis and modification in chondrocytes. Nature. 2019;565:511–5.
Xie M, Tang Q, Nie J, Zhang C, Zhou X, Yu S, et al. BMAL1-downregulation aggravates porphyromonas gingivalis-induced atherosclerosis by encouraging oxidative stress. Circ Res. 2020;126:e15–e29.
Acknowledgements
We would like to thank Prof. Anbing Shi (Huazhong University of Science and Technology) for constructive advice. We also thanks Prof. Ying Xu (Soochow University) for the Bmal1wt/− and Bmal1fl/fl mating pairs.
Funding
This research was funded by the National Natural Science Foundation of China for Key Program Projects (82030070, to LC) and General Program (82270950, to QT), Hubei Provincial Natural Science Fund for Creative Research Group (2020CFA014, to LC), Young Talent Program by Health Commission of Hubei Province (WJ2021Q059, to QT) and the Youth Clinical Research Fund of Chinese Stomatological Association (CSA-O2020-10, to QT).
Author information
Authors and Affiliations
Contributions
GC, QT and LC performed the experiments, data analysis and drafted the manuscript. SY, JS and YS contributed to the materials and analysis methods. GF, GM, YZ, YY, JP and XL contributed to animal housing and animal experiments. QW and LZ supervised the mathematical aspects of this research. LC supervised the whole study and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
All experiments were approved by the Institutional Animal Care and Use Committee of Tongji Medical College (LAUCU Number:2809).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, G., Tang, Q., Yu, S. et al. Developmental growth plate cartilage formation suppressed by artificial light at night via inhibiting BMAL1-driven collagen hydroxylation. Cell Death Differ 30, 1503–1516 (2023). https://doi.org/10.1038/s41418-023-01152-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01152-x
- Springer Nature Limited
This article is cited by
-
Time of exercise differentially impacts bone growth in mice
Nature Metabolism (2024)