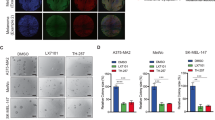
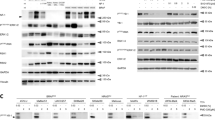
Abstract
RAC1P29S is the third most prevalent hotspot mutation in sun-exposed melanoma. RAC1 alterations in cancer are correlated with poor prognosis, resistance to standard chemotherapy, and insensitivity to targeted inhibitors. Although RAC1P29S mutations in melanoma and RAC1 alterations in several other cancers are increasingly evident, the RAC1-driven biological mechanisms contributing to tumorigenesis remain unclear. Lack of rigorous signaling analysis has prevented identification of alternative therapeutic targets for RAC1P29S-harboring melanomas. To investigate the RAC1P29S-driven effect on downstream molecular signaling pathways, we generated an inducible RAC1P29S expression melanocytic cell line and performed RNA-sequencing (RNA-seq) coupled with multiplexed kinase inhibitor beads and mass spectrometry (MIBs/MS) to establish enriched pathways from the genomic to proteomic level. Our proteogenomic analysis identified CDK9 as a potential new and specific target in RAC1P29S-mutant melanoma cells. In vitro, CDK9 inhibition impeded the proliferation of in RAC1P29S-mutant melanoma cells and increased surface expression of PD-L1 and MHC Class I proteins. In vivo, combining CDK9 inhibition with anti-PD-1 immune checkpoint blockade significantly inhibited tumor growth only in melanomas that expressed the RAC1P29S mutation. Collectively, these results establish CDK9 as a novel target in RAC1-driven melanoma that can further sensitize the tumor to anti-PD-1 immunotherapy.








Similar content being viewed by others
Data availability
All raw sequencing data has been deposited in the Gene Expression Omnibus database under accession #GSE248218. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [49] partner repository with the dataset identifier PXD046960. All other raw data generated from this study are available from the corresponding author upon request.
References
Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell 2012;150:251–63.
Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14.
Davis MJ, Ha BH, Holman EC, Halaban R, Schlessinger J, Boggon TJ. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc Natl Acad Sci USA. 2013;110:912–7.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl J Med. 2011;364:2507–16.
Giugliano F, Crimini E, Tarantino P, Zagami P, Uliano J, Corti C, et al. First line treatment of BRAF mutated advanced melanoma: Does one size fit all? Cancer Treat Rev. 2021;99:102253.
Watson IR, Li L, Cabeceiras PK, Mahdavi M, Gutschner T, Genovese G, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–52.
Vu HL, Rosenbaum S, Purwin TJ, Davies MA, Aplin AE. RAC1 P29S regulates PD-L1 expression in melanoma. Pigment Cell Melanoma Res. 2015;28:590–8.
Lodde GC, Jansen P, Herbst R, Terheyden P, Utikal J, Pfohler C, et al. Characterisation and outcome of RAC1 mutated melanoma. Eur J Cancer. 2023;183:1–10.
Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004.
Moore KA, Sethi R, Doanes AM, Johnson TM, Pracyk JB, Kirby M, et al. Rac1 is required for cell proliferation and G2/M progression. Biochem J. 1997;326:17–20.
Michaelson D, Abidi W, Guardavaccaro D, Zhou M, Ahearn I, Pagano M, et al. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J Cell Biol. 2008;181:485–96.
Maroto B, Ye MB, von Lohneysen K, Schnelzer A, Knaus UG. P21-activated kinase is required for mitotic progression and regulates Plk1. Oncogene. 2008;27:4900–8.
Zhao ZS, Lim JP, Ng YW, Lim L, Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol Cell. 2005;20:237–49.
Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–83.
Anshabo AT, Milne R, Wang S, Albrecht H. CDK9: A Comprehensive Review of Its Biology, and Its Role as a Potential Target for Anti-Cancer Agents. Front Oncol. 2021;11:678559.
Guhan SM, Shaughnessy M, Rajadurai A, Taylor M, Kumar R, Ji Z, et al. The Molecular Context of Vulnerability for CDK9 Suppression in Triple Wild-Type Melanoma. J Invest Dermatol. 2021;141:2018–27.e4.
Itzen F, Greifenberg AK, Bosken CA, Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42:7577–90.
Zhang H, Pandey S, Travers M, Sun H, Morton G, Madzo J, et al. Targeting CDK9 Reactivates Epigenetically Silenced Genes in Cancer. Cell. 2018;175:1244–58.e26.
Hossain DMS, Javaid S, Cai M, Zhang C, Sawant A, Hinton M, et al. Dinaciclib induces immunogenic cell death and enhances anti-PD1-mediated tumor suppression. J Clin Invest. 2018;128:644–54.
Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548:471–5.
Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 2018;175:984–97.e24.
Deng J, Wang ES, Jenkins RW, Li S, Dries R, Yates K, et al. CDK4/6 Inhibition Augments Antitumor Immunity by Enhancing T-cell Activation. Cancer Discov. 2018;8:216–33.
Schaer DA, Beckmann RP, Dempsey JA, Huber L, Forest A, Amaladas N, et al. The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep. 2018;22:2978–94.
Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–5.
Adams AB, Ford ML, Larsen CP. Costimulation Blockade in Autoimmunity and Transplantation: The CD28 Pathway. J Immunol. 2016;197:2045–50.
Mohan AS, Dean KM, Isogai T, Kasitinon SY, Murali VS, Roudot P, et al. Enhanced Dendritic Actin Network Formation in Extended Lamellipodia Drives Proliferation in Growth-Challenged Rac1(P29S) Melanoma Cells. Dev Cell. 2019;49:444–60.e9.
Lionarons DA, Hancock DC, Rana S, East P, Moore C, Murillo MM, et al. RAC1(P29S) Induces a Mesenchymal Phenotypic Switch via Serum Response Factor to Promote Melanoma Development and Therapy Resistance. Cancer Cell. 2019;36:68–83.e9.
Gorgun FM, Widen SG, Tyler DS, Englander EW. Enhanced Antitumor Response to Immune Checkpoint Blockade Exerted by Cisplatin-Induced Mutagenesis in a Murine Melanoma Model. Front Oncol. 2021;11:701968.
Ribas A, Lawrence D, Atkinson V, Agarwal S, Miller WH Jr., Carlino MS, et al. Combined BRAF and MEK inhibition with PD-1 blockade immunotherapy in BRAF-mutant melanoma. Nat Med. 2019;25:936–40.
Zhang B, Zhong X, Sauane M, Zhao Y, Zheng ZL. Modulation of the Pol II CTD Phosphorylation Code by Rac1 and Cdc42 Small GTPases in Cultured Human. Cells. 2020;9:621.
Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–31.
Vollmuth F, Blankenfeldt W, Geyer M. Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J Biol Chem. 2009;284:36547–56.
Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–909.
Du Z, Song X, Yan F, Wang J, Zhao Y, Liu S. Genome-wide transcriptional analysis of BRD4-regulated genes and pathways in human glioma U251 cells. Int J Oncol. 2018;52:1415–26.
Li L, Meng Y, Wu X, Li J, Sun Y. Bromodomain-containing protein 4 inhibitor JQ1 promotes melanoma cell apoptosis by regulating mitochondrial dynamics. Cancer Sci. 2021;112:4013–25.
Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013;73:6264–76.
Dar AA, Bezrookove V, Nosrati M, Ice R, Patino JM, Vaquero EM, et al. Bromodomain inhibition overcomes treatment resistance in distinct molecular subtypes of melanoma. Proc Natl Acad Sci USA. 2022;119:e2206824119.
Echevarria-Vargas IM, Reyes-Uribe PI, Guterres AN, Yin X, Kossenkov AV, Liu Q, et al. Co-targeting BET and MEK as salvage therapy for MAPK and checkpoint inhibitor-resistant melanoma. EMBO Mol. Medi. 2018;10:e8446.
Nikbakht N, Tiago M, Erkes DA, Chervoneva I, Aplin AE. BET Inhibition Modifies Melanoma Infiltrating T Cells and Enhances Response to PD-L1 Blockade. J Invest Dermatol. 2019;139:1612–5.
Kagoya Y, Nakatsugawa M, Yamashita Y, Ochi T, Guo T, Anczurowski M, et al. BET bromodomain inhibition enhances T cell persistence and function in adoptive immunotherapy models. J Clin Invest. 2016;126:3479–94.
Shibata H, Saito S, Uppaluri R. Immunotherapy for Head and Neck Cancer: A Paradigm Shift From Induction Chemotherapy to Neoadjuvant Immunotherapy. Front Oncol. 2021;11:727433.
Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16:2598–608.
Mandal R, Senbabaoglu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1:e89829.
Senyuz S, Jang H, Nussinov R, Keskin O, Gursoy A. Mechanistic Differences of Activation of Rac1(P29S) and Rac1(A159V). J Phys Chem B. 2021;125:3790–802.
Storch K, Cordes N. The impact of CDK9 on radiosensitivity, DNA damage repair and cell cycling of HNSCC cancer cells. Int J Oncol. 2016;48:191–8.
Skvortsov S, Dudas J, Eichberger P, Witsch-Baumgartner M, Loeffler-Ragg J, Pritz C, et al. Rac1 as a potential therapeutic target for chemo-radioresistant head and neck squamous cell carcinomas (HNSCC). Br J Cancer. 2014;110:2677–87.
Perez-Riverol Y, Bai J, Bandla C, Garcia-Seisdedos D, Hewapathirana S, Kamatchinathan S, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D52.
Acknowledgements
We would like to thank Ruth Halaban and Gaudenz Danuser for cell lines. We would also like to thank Fox Chase Cancer Center core facilities including flow cytometry, biological imaging, histopathology, laboratory animal and biostatistics facilities. This work was supported by grants from the NIH R01 CA2271844 (JC), the Melanoma Research Foundation (JC), the F.M. Kirby Foundation (CUA), NIH R01 CA211670 (JSD), NIH T32 CA009035 (AMK), and NCI Core Grant P30 CA06927 (to Fox Chase Cancer Center).
Author information
Authors and Affiliations
Contributions
AC: Conceptualization, data curation, formal analysis, investigation, methodology, project administration, supervision, visualization, writing – original draft, writing – review and editing. KB: Conceptualization, formal analysis, investigation, writing – review and editing. CU-A: Funding acquisition, methodology, writing – review and editing. AK: Formal analysis, investigation, software. DA-O: Methodology QC: Formal analysis, investigation, methodology, software. SP: Data Curation, formal analysis, software. YZ: Data Curation, formal analysis, software JD: Methodology, resources, supervision, formal analysis. JC: Conceptualization, funding acquisition, methodology, project administration, resources, supervision, visualization, writing – original draft, writing – review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cannon, A.C., Budagyan, K., Uribe-Alvarez, C. et al. Unique vulnerability of RAC1-mutant melanoma to combined inhibition of CDK9 and immune checkpoints. Oncogene 43, 729–743 (2024). https://doi.org/10.1038/s41388-024-02947-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-024-02947-z
- Springer Nature Limited