Abstract
Enduring loneliness is associated with mental disorders and physical diseases. Although genome-wide association studies (GWAS) have identified risk loci associated with loneliness, how these loci confer the risk remains largely unknown. In the current study, we aimed to investigate key proteins underlying loneliness in the brain by integrating human brain proteomes and transcriptomes with loneliness GWAS to perform a discovery proteome-wide association study (PWAS), followed by a confirmatory PWAS, transcriptome-wide association analysis (TWAS), Mendelian randomization (MR), Steigering filtering analysis and Bayesian colocalization analysis. Moreover, given the fact that loneliness is associated with mental disorders, we explored the shared genetic architecture between loneliness and mental disorders. Totally, we identified 18 genes to be associated with loneliness via their cis-regulated brain protein abundance. Eleven of the 18 genes (61.1%) were replicated in the confirmatory PWAS, and mRNA levels of 4 genes were further validated to be associated with loneliness.MR and genetic colocalization analysis further confirmed that the increased protein abundance of ALDH2 and ICA1L was protective against loneliness, while the increased protein abundance of GPX1 was a risk for developing loneliness. Furthermore, we found genetic correlations, bidirectional causal associations and overlapping phenotype-associated protein profiles between loneliness and mental disorders including major depression and schizophrenia. In summary, our findings provided clues about the brain-related molecular basis underlying loneliness, which warrants further investigation.
Similar content being viewed by others
Introduction
Human beings are inherently social, and social relationships are important in maintaining both our physical and mental health [1, 2]. However, enduring loneliness, whose core experience consists of social isolation and the absence of both rational and collective connectedness [3, 4], is associated with an increased risk of mental disorders (including depression, schizophrenia, anxiety, suicide, and dementia), physical diseases (such as cardiovascular disease and diabetes), and all-cause mortality [5,6,7,8,9,10,11]. Therefore, it is important to decipher the etiology and mechanism underlying loneliness, which can provide insights into reducing its related morbidity and mortality. Although a wide range of environmental, social, economic, and behavioral factors have been identified to contribute to the occurrence of loneliness [12], the genetic factor has also been found to play an important role. On the one hand, twin studies have found that the heritability of loneliness is about 40–50% [13]. On the other hand, genome-wide association studies (GWASs) have also identified several genetic variants associated with loneliness [9, 14]. However, it remains unclear how these genetic loci involve in the loneliness-related biological processes, which hampers further understanding of the mechanisms underlying loneliness.
Proteins are the main function molecules in the body which perform a vast array of functions [15], while little is known about which proteins in the central nervous system are responsible for loneliness. In the current study, we aimed to investigate the crucial proteins underlying loneliness in the brain with an integrative analytic approach. Firstly, we integrated 2 protein quantitative trait loci (pQTL) datasets obtained from the human brain and the GWAS summary statistics for loneliness to perform proteome-wide association study (PWAS) to identify proteins associated with loneliness. Next, we further validated the association of the significant proteins at the transcriptional level by transcriptome-wide association analysis (TWAS). Thirdly, we applied 2-sample Mendelian randomization (MR) analysis and Steigering filtering analysis to confirm the causal associations of significant proteins. Then, we applied genetic colocalization analysis to assess the probability that two traits share the same causal variant. Moreover, we further validated the causal associations at the transcriptional level. Lastly, given the facts that loneliness is associated with an increased risk of mental disorders, and there is a high prevalence of loneliness in mental disorders [16], we investigated the genetic correlation, bidirectional causal associations and shared phenotype-associated protein profiles between loneliness and mental disorders, including major depression, bipolar disorder and schizophrenia.
Methods
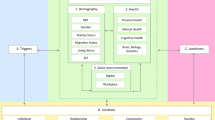
The flowchart of the study was presented in Fig. 1.
Datasets
The summary of the datasets used in the current study were listed in Supplementary Table 1.
Human brain proteome reference used in the PWAS
In the discovery phase, the human brain proteome was profiled from the dorsal lateral prefrontal cortex(dlPFC) of 376 participants of the European Ancestry of the Religious Orders Study/Memory and Aging Project (ROS/MAP) cohorts [17]. To obtain the genetic profile of protein abundance: firstly, protein abundance was measured and genotyping was performed; then quality control was performed and confounding factors such as demographic characteristics of the participants and technical bias were corrected; finally, genotypes and protein abundance were integrated to estimate the genetic effects on protein abundance, which is referred to protein weights. 8356 proteins were included in the proteomic profiles, of which 1475 were heritable and their protein weights were used in the PWAS [18]. The detailed methods can be found in the original study [18]. In the confirmatory phase, the human brain proteome was derived from the dlPFC of another cohort of European individuals recruited by the Banner Sun Health Research Institute. The process of proteome genetic profiling was similar to that in the discovery PWAS. After quality control, there were 152 individuals with both proteomic and genetic data to include in the confirmation TWAS [18].
Human brain transcriptome reference used in the TWAS
The Human brain transcriptome reference used in the TWAS was derived from the dlPFC of 452 European individuals recruited by the CommonMind Consortium (CMC). The detailed methods for genetic profiling were described by the original study [19]. 5420 mRNA levels were found to be heritable and their mRNA weights can be downloaded at http://gusevlab.org/projects/fusion/.
Human brain pQTL datasets used in the 2-sample MR
The Human brain-tissue-derived pQTL dataset used in the 2-sample MR was the same as the one used in the discovery PWAS, which was derived from 376 participants of the ROS/MAP cohort. However, in MR, we selected eligible instrument variables (IVs) at a stringent genome-wide significant threshold at p < 5E-08 [18]. Totally, 607 proteins were found to have significantly associated IVs with p < 5E-08.
Human brain eQTL datasets used in the 2-sample MR
The brain eQTL dataset was obtained from the PsychENCODE Consortium which obtained data from the human brain over a sample size of 1866 individuals of European Ancestry [20]. After filtering, there were 10,489 genes in dlPFC that have significantly associated IVs with p < 5E-08.
GWAS summary statistics for loneliness
The dataset for loneliness was derived from the GWAS performed in UK Biobank study participants [14]. Briefly, three related questions assessing loneliness and social isolation were used—(1) “Do you often feel lonely”?, to which individuals answered “yes” (recorded as cases) or “no” (controls), (2) A composite variable based on the questions “Including yourself, how many people are living together in your household?” and “How often do you visit friends or family or have them visit you?” (cases were defined as those who lived alone and who indicated that they either never visited or had no friends or family outside their household; controls were defined as those who either did not live alone, or had friends who visited at least once a week) and (3) A variable representing the quality of social interactions “How often are you able to confide in someone close to you?” (cases were defined as those who answered “Never or almost never”, controls were defined as those who answered “Almost daily”). And GWAS was performed independently based on each trait. Then, results from these three GWAS were combined using multi-trait GWAS (MTAG) into a single discovery sample, yielding an effective sample size of 487,647 individuals [14]. Totally, 15 genomic loci (p < 5E-08) were identified to be associated with loneliness [14].
GWAS summary statistics for major depression, bipolar disorder and schizophrenia
The GWAS summary datasets of mental disorders including major depression, schizophrenia and bipolar disorder were obtained from the latest and largest GWASs performed by Psychiatric Genomic Consortium (PGC) in the European population. Briefly, the summary statistics of major depression originated from a meta-analysis of PGC and UK Biobank datasets, which included 170,756 cases and 329,443 control [21], The dataset of bipolar disorder was from the third GWAS meta-analysis of the PGC Bipolar Disorder Working Group, comprising 41,917 individuals with BD and 371,549 controls [22]. The dataset of schizophrenia was from the most recent and largest GWAS, which included 53,386 cases and 77,258 control [23].
Statistical analyses
PWAS and TWAS
FUSION is a powerful strategy integrating gene expression/protein abundance measurements with the summary statistics from GWAS to identify genes whose cis-regulated expression/protein abundance is associated with complex traits [24]. We applied the FUSION pipeline according to the author’s guideline with default settings to perform TWAS and PWAS [24]. The major histocompatibility complex (MHC) region on chromosome 6 was excluded from the analysis because of its structural complexity. False discovery rate (FDR) was applied in the multiple testing.
MR analysis
MR is a genetic method using the genome-wide significant single nucleotide polymorphisms (SNPs) that were strongly associated with the exposure as IVs, to investigate the causal link between exposure and outcome [25]. It has been widely used in exploring etiologies of complex traits. The most important and fundamental step of MR is to include eligible IVs [25]. To identify eligible IVs, three key assumptions must be met [25]. Assumption 1 is the relevance assumption, which requires the genetic variant to be directly associated with the exposure. To meet assumption 1, we restricted the SNPs to be directly associated with the exposure at p < 5E-08 (genome-wide significant threshold). Assumption 2 is the independence assumption, which requires that the genetic variant should not be directly related to confounding factors. And assumption 3 is the exclusion assumption, which requires that the genetic variant should not be directly associated with the outcome. Both assumption 2 and assumption 3 can be calculated as horizontal pleiotropy in the post-MR analysis [25].
Once eligible IVs were selected, they were linkage disequilibrium (LD) clumped with r2 < 0.001 in a 10-megabase distance. Next, the IVs were extracted from the outcome trait and were harmonized in both exposure and outcome GWAS datasets. Then, MR effects were estimated by the Wald ratio if only a single independent IV was available, and by the inverse variance weighted (IVW) method if two or more IVs were available [26]. As a result, p < 0.05 was considered as suggestively significant. Next, to ensure the causation was not distorted by the presence of reverse causation, the Steiger filtering method was applied [27], and p < 0.05 indicated that the effect direction is from the exposure to the outcome. Moreover, when 3 or more IVs were available, sensitivity analyses including MR-Egger, weighted mode, weighted median mode and simple mode analysis were performed to ensure the robustness of the results [26], and post-MR analyses including MR Egger intercept test for horizontal pleiotropy, Cochran’s Q test for heterogeneity, and MR-PRESSO for outlier test were also performed [25]. However, after clumping, there were 1 or 2 IVs for most of the pQTLs, therefore, sensitivity analyses and post-MR analyses were unable to be performed. The above-mentioned algorithms were implemented with the “TwoSampleMR” R package (github.com/MRCIEU/TwoSampleMR) [7].
Bayesian colocalization analysis
Bayesian colocalization analysis was performed to assess whether two association signals are consistent with a shared causal variant [28]. The analysis was conducted with COLOC in the R package with default parameters, which tested the posterior probability of 5 hypotheses: H0: No association with either trait; H1: Association with trait 1, not with trait 2; H2: Association with trait 2, not with trait 1; H3: Association with trait 1 and trait 2, two independent SNPs; H4: Association with trait 1 and trait 2, one shared SNP [28]. And a posterior probability of Hypothesis 4 (PPH4) > 0.8 was considered as that the two association signals are consistent with a shared causal variant [28].
Cell-type specific expression of the loneliness-related genes
The cell-type-specific expression profile of the potential causal genes in the brain was downloaded from the human single-cell RNA-seq data from the Cell Types Database (https://portal.brain-map.org/atlases-and-data/rnaseq). Cell-type expression specificity (CELLEX), a tool for computing cell-type Expression Specificity (ES) profiles, was applied to capture multiple aspects of ES [29].
Cross-trait LDSC between loneliness and mental disorders
GWAS summary statistics of loneliness, major depression, schizophrenia and bipolar disorder were obtained as previously described. We assessed the genetic correlation between loneliness and mental disorders with the LD Score Regression software (v1.0.0) (https://github.com/bulik/ldsc) [30].
Shared phenotype-associated proteins between loneliness and mental disorders
To further explore the shared genetic basis between loneliness and mental disorders, we compared the shared significant phenotype-associated proteins (FDR < 0.05) between loneliness and mental disorders by performing PWAS on loneliness and mental disorders using the ROSMAP brain proteome reference panel.
Results
Discovery and confirmatory PWAS for loneliness
In the discovery PWAS, we identified 18 genes whose cis-regulated protein levels in the brain were associated with the risk of developing loneliness with an FDR < 0.05 (Table 1; Fig. 2A; Supplementary Table 2). Among these 18 genes, 2 are located within 500 kb of each other on chromosome 20 (CSE1L and STAU1). The conditional analysis identified CSE1L in the pair as jointly significant, while STAU1 was no longer significant and was dropped out. Then, we performed an independent confirmatory PWAS using a different reference brain proteome to confirm our findings. 11 (61.1%) genes were replicated. The remaining 7 genes not replicated were not mapped in confirmatory PWAS, which might be caused by the relatively small sample size (Table 1; Fig. 2A; Supplementary Table 3).
A Manhattan plot for the loneliness PWAS integrating the loneliness GWAS with the discovery ROS/MAP proteomes (N = 376) and the confirmatory Banner proteome (N = 152). Each point represents a single association test between a gene and loneliness ordered by genomic position on the x-axis and the association strength on the y-axis as the −log10(P) of a z-score test. The discovery of PWAS identified 18 genes whose cis-regulated brain protein abundance was associated with loneliness at an FDR of P < 0.05, 11 of which were replicated in the confirmatory PWAS (blue labeled). The red horizontal line reflects the significant threshold of the FDR P < 0.05 and is set at the highest unadjusted p-value that is below that threshold (p = 7 × 10 − 4). B Forest plot of the MR-identified genes underlying loneliness; blue triangle: protein level; black rhombus: transcriptional level; WR wald ratio. C The overlapping phenotype-associated proteins between loneliness and mental disorders; white block: not overlapping between loneliness and the disease; green block: overlapping between loneliness and the disease, and with the same direction of effect; orange block: overlapping between loneliness and the disease, and with the opposite direction of effect; BPD bipolar disorder, SCZ schizophrenia.
Confirmation of the loneliness-associated genes at the transcriptional level
Given the fact that proteins were translated by mRNA, we aimed to explore the loneliness-associated genes at the mRNA level. To this end, we performed a TWAS using CMC reference brain transcriptomes. We found that cis-regulated brain mRNA levels of 48 genes were found to be associated with loneliness. Four of the 11 loneliness-associated genes identified by the PWAS were validated at the transcriptional level, including CSE1L, ALDH2, GPX1, and ICA1L. Notably, the transcriptional level and protein level of ALDH2 and GPX1 had the same direction of effect on loneliness, while ICA1L had opposite directions of effect on loneliness between transcription level and protein level (Table 1; Supplementary Table 4).
MR and colocalization verified 3 genes having causal associations with loneliness
To further confirm the causal associations between brain proteome and loneliness, we further applied a 2-sample MR analysis. Among the 11 proteins discovered to be associated with loneliness, 4 of which were found to have causal associations with loneliness by the Wald ratio method (Table 1; Supplementary Table 5). Specifically, the increased abundance of TMEM106B and GPX1 led to an increased loneliness risk, while the increased abundance of ALDH2 and ICA1L led to a decreased loneliness risk. Steiger filtering analysis confirmed the correct direction of causal associations. Furthermore, colocalization analysis revealed that 3 out of the 4 genes (ALDH2, ICA1L, and GPX1) offered evidence of genetic colocalization. Moreover, we further confirmed the causal associations of ALDH2, ICA1L, and GPX1 at the transcriptional level. Notably, the mRNA level of ALDH2 and GPX1 have the same direction of effect on loneliness as the protein level, while the mRNA level of ICA1L has the opposite direction of effect on loneliness as compared to the protein level (Table 1; Fig. 2B).
Summary findings
According to the results of PWAS, TWAS, MR, and colocalization, we assigned levels of confidence for the loneliness-related genes. We had very high confidence in ALDH2, ICA1L, and GPX1, because PWAS, TWAS, MR, and colocalization results all support their causal associations with loneliness. We also had high confidence in CSE1L, because both PWAS, TWAS and transcriptional MR analysis all supported its role in loneliness, while it was not profiled in the protein MR because it did not have IV with p < 5E-08. However, we had moderate confidence in TMEM106B, STXBP6, ORC4, PACSIN3, TAOK3, CTNND1, and CNTN2. Although they were both confirmed by an independent PWAS, they were not supported at the transcriptional level as well as genetic colocalization, which indicated that the signals discovered by PWAS might arise from linkage disequilibrium (LD, i.e., SNP affecting protein abundance is in LD with SNP affecting loneliness) or pleiotropy (SNP affecting both protein abundance and loneliness). Furthermore, we had low confidence in PDIA3, PACSIN1, EPHB2, TMEM25, DCUN1D1, and PLCD3, because they were not confirmed by the confirmatory PWAS (Table 2).
Cell-type specific expression of the loneliness-related genes
We found that ICA1L and ALDH2 had enrichment in different neurons. Specifically, ICA1L was enriched in Glutaminergic neurons, GABAergic neurons, and astrocytes, and ALDH2 was enriched in astrocytes and GABAergic neurons (Supplementary Table 6).
Loneliness and mental disorders have a shared genetic profile
With LDSC, we observed a positive genetic correlation between loneliness and mental disorders, including depression (rg = 0.582, p = 6.55E-163), schizophrenia (rg = 0.18, p = 5.87E-18) and bipolar disorder (rg = 0.118, p = 2.11E-06), suggesting that there were 58.2% shared genetic basis between loneliness and major depression, 18% shared genetic basis between loneliness and schizophrenia, and 11.8% shared genetic basis between loneliness and bipolar disorder.
Naturally, we wondered about the direction of causal associations between loneliness and these mental disorders, therefore, we performed a bidirectional MR analysis. On the one hand, in the forward MR analysis (loneliness as exposure and mental disorders as outcomes), we found that loneliness was contributing to an increased risk of developing major depression (ORIVW [95% CI] = 2.14 [1.45–3.16], pIVW = 1.38E-04) and schizophrenia (ORIVW [95% CI] = 3.53[1.48–8.41], pIVW = 0.004) by the IVW method, which is further confirmed by other sensitivity analyses. However, loneliness was not associated with the risk of developing bipolar disorder (ORIVW [95% CI] = 2.00 [0.90–4.43], pIVW = 0.089) by the IVW method, as well as by other sensitivity analyses (Table 3). On the other hand, in the reverse MR analysis (mental disorders as exposures and loneliness as outcome), we found that major depression was strongly associated with the risk of developing loneliness (ORIVW [95% CI] = 1.17 [1.13–1.22], pIVW = 3.50E-16), which was also supported by other sensitivity analyses. While schizophrenia was marginally associated with the risk of developing loneliness (ORIVW [95% CI] = 1.02 [1.01–1.03], pIVW = 1.27E-05), which was supported by weighted median mode. However, bipolar disorder was not associated with the risk of loneliness (ORIVW [95% CI] = 1.00 [0.99–1.02], pIVW = 0.623) (Table 3). Although some heterogeneity was detected among the IVs, no horizontal pleiotropy and outlier was detected in all the analyses (Supplementary Table 7).
Moreover, to further explore the shared genetic basis between loneliness and mental disorders, we compared the shared phenotype-associated proteins (FDR < 0.05) between loneliness and mental disorders. Among the 18 loneliness-associated proteins in the brain, 5 (5/18, 27.8%) proteins (namely TMEM106B, TAOK3, CTNND1, EPHB2 and TMEM25) were found to be associated with major depression, all of which have the same direction of effect on loneliness and major depression. Moreover, 6 proteins (6/18, 33.3%) were found to be also associated with schizophrenia, with 5 proteins (namely CSE1L, CTNND1, CNTN2, STAU1 and TMEM25) having the same direction of effect on loneliness and schizophrenia, while PACSIN3 had the opposite direction of effect on loneliness and schizophrenia. Furthermore, 8 proteins (8/18, 44.4%) were found to also be associated with bipolar disorder. However, only 2 proteins (namely TMEM106B and PLCD1) had the same direction of effect on loneliness and bipolar disorder (Fig. 2C; Supplementary Table 8).
Discussion
In the current study, we applied a discovery and confirmatory study design, and integrated the PWAS, TWAS, MR, and colocalization, aiming to discover the genetically-determined protein levels associated with loneliness in the brain. We identified 18 proteins to be associated with loneliness in the discovery PWAS, 11 (64.7%) of them were replicated in the confirmatory PWAS, and 4 out of the 11 genes were further confirmed by TWAS, providing an additional layer of confirmation. Notably, the causal associations of 3 genes (ALDH2, GPX1, and ICA1L) were further validated by MR and genetic colocalization. Last but not least, we observed a significant genetic correlation and shared genetic profile between loneliness and mental disorder, especially major depression and schizophrenia.
Specifically, we have high confidence that the increased protein level of ALDH2 and ICA1L in the brain was contributing to a lower risk of developing loneliness. The protective role of ALDH2 towards loneliness could be explained by the acetate-GABA pathway. ALDH2 (aldehyde dehydrogenase) is a key enzyme in ethanol metabolism, catalyzing acetaldehyde’s conversion into acetate [31]. Acetate has been found to promote the synthesis of Gamma-Aminobutyric Acid(GABA) [32]. GABA is an important inhibitor neurotransmitter, which blocks certain nervous signals to reduce fear, anxiety, and stress [33]. A decreased GABA function in the brain is associated with panic disorder [34], anxiety [35], and decreased sociability [36]. Therefore, the increased abundance of ALDH2 might lead to an increased level of acetate and GABA, which might decrease the risk of developing loneliness. However, further functional studies are warranted to understand the role of acetate and GABA in loneliness.
ICA1L was also found to be protective against loneliness. ICA1L is abundantly expressed in the testes and the brain, it is crucial for acrosome, nucleus, and mitochondrial sheath formation [37], while its role in CNS remains unclear. Moreover, it has been found to decrease the risk for Alzheimer’s disease(AD) [18, 38] and small vessel disease(SVD) [39, 40]. Notably, in our analysis, we found that ICA1L had opposite effect directions on loneliness between mRNA level and protein level. Specifically, both in the TWAS and transcriptional level MR, we found that an increased mRNA level of ICA1L was associated with a higher risk of loneliness. While in the PWAS and protein level MR, we found that an increased protein level of ICA1L was associated with a lower risk of loneliness. These inconsistent roles between mRNA and protein levels of ICA1L might be caused by post-transcriptional modifications such as mRNA splicing and protein degradation [41]. Together with our results, these findings suggested that ICA1L is important in CNS-related disorders, while further studies were needed to ascertain its role in loneliness.
On the contrary, the increased abundance of GPX1 was associated with a higher risk of developing loneliness. GPX1 (Glutathione peroxidase) is a selenium-containing enzyme functioning in the reduction of hydrogen peroxide and alkyl hyperoxides and was identified to be a risk locus for loneliness by the original GWAS [14]. It was found to be protective against trauma-induced brain injury [42] and brain ischemia-reperfusion injury [43]. However, its contribution to loneliness needs further exploration.
In the cell-specific expression analysis, we found that both ICA1L and ALDH2 were enriched in GABAergic neurons and astrocytes. As we discussed before, GABA is important in dealing with fear, anxiety, and stress [33]. Moreover, astrocytes have been found to actively interact with local neural circuits and play a crucial role in information processing and generating physiological and behavioral responses. These results indicated that GABAergic neurons and astrocytes should get more attention in social interaction [44].
Our study also explored the shared genetic basis between loneliness and mental disorders. Firstly, we observed a high genetic correlation (rg = 0.58), bidirectional causal associations and 5 shared phenotype-associated proteins between loneliness and major depression. These results were consistent with previous findings, which also identified a strong genetic correlation and shared loci between loneliness and depression [45, 46]. Together with our findings, these results indicated a strong role of genetic influences in the co-occurrence of loneliness and depression. And we also observed a significant genetic correlation between loneliness and schizophrenia at rg = 0.18, comparable to the previous studies that used the PGC2.0 schizophrenia dataset and observed a genetic correlation between loneliness and schizophrenia at rg = 0.17 [14]. Moreover, we also observed bidirectional causal associations between loneliness and schizophrenia, with a greater effect size of loneliness on schizophrenia than in the opposite direction, which is consistent with the results from a previous study which identified that the loneliness polygenic score was contributing to schizophrenia risk, as well as a bidirectional causal relationship between loneliness and schizophrenia [47]. The causal association between loneliness and schizophrenia could explain that loneliness and objective social isolation could trigger both positive and negative psychotic symptoms [48], and could also explain the high levels of loneliness and isolation before the onset of psychosis in individuals at clinical high risk for psychosis [16]. On the other hand, the causal association between schizophrenia and social isolation could also explain the high prevalence of loneliness in the chronic stages of psychotic illnesses [48, 49]. Accordingly, in the current study, we also observed 6 overlapped protein signals between loneliness and schizophrenia in the PWAS study, most of which had the same direction of effects. In brief, our study provided additional evidence for supporting the role of loneliness in the development of schizophrenia. However, we found a weaker genetic correlation between loneliness and bipolar disorder and failed to identify the causal association between loneliness and bipolar disorder. Moreover, we found that although there were 8 phenotype-associated proteins between bipolar disorder and loneliness, most of which had opposite directions of effect. These results suggested that loneliness and bipolar disorder might have different mechanisms, because patients with bipolar disorder not only have depressive symptoms but also have elevated or irritable moods [50]. In general, our study provided supplementary evidence about the shared genetic profile between loneliness and mental disorders, which is consistent with a previous study that identified 149 loci jointly associated with loneliness and mental disorders (major depression nloci = 67, schizophrenia nloci = 54, and bipolar disorder nloci = 28) [11]. Given that loneliness is modifiable, it could be an effective target for preventing its effects on mental health, especially major depression and schizophrenia.
The current study has some strengths. First, we used discovery and validation design to enhance the level of confidence in our findings. Second, to the best of our knowledge, this is the first study that performed both PWAS, TWAS, protein-level MR and mRNA-level MR to gain more insights into the action of genes at both the protein level and transcriptional level. Thirdly, we performed MR and Bayesian colocalization analysis to further confirm the causal associations of the brain proteins. Last but not least, we provided additional evidence supporting the shared genetic architecture of loneliness and mental disorders, especially major depression and schizophrenia.
There were also some limitations that should be acknowledged in the current study. Firstly, the SNP-based heritability of the brain proteome is limited which might be due to the relatively small sample size and the limited number of reference brain proteomes in the original studies [18]. Future studies with a larger number of human proteomes can have more power to identify heritable proteins. Besides, all evidence presented in the current study was obtained from statistical analyses, while the mechanisms of how those candidate genes contribute to loneliness remain unclear, and further functional studies are needed to validate our findings. Moreover, the measurement of loneliness in the UK Biobank is not comprehensive, and a more comprehensive assessment of loneliness would be beneficial. Last but not least, the datasets used in the present analysis were derived from the European population, confirmatory studies are needed in other ethnicities.
Conclusion
In summary, we provided multiple levels of evidence to support the causal associations between 3 brain proteins (ALDH2, ICA1L, and GPX1) and loneliness. Moreover, we provided additional evidence supporting the shared genetic architecture between loneliness and mental disorders including major depression and schizophrenia. In general, our findings provided clues about the brain-related molecular basis underlying loneliness, which warrants further investigation.
Data availability
The data were obtained from the publicly available database (listed in Supplementary Table 1). The datasets analyzed during the current study are available from the corresponding author upon reasonable request.
References
Umberson D, Montez JK. Social relationships and health: a flashpoint for health policy. J Heal Soc Behav. 2010;51:S54–66.
HJ S, LK R, Debra U. Social Relationships and Health. Science (80-). 1988;241:540–5.
Russell D, Peplau LA, Cutrona CE. The revised UCLA Loneliness Scale: concurrent and discriminant validity evidence. J Pers Soc Psychol. 1980;39:472–80.
Cacioppo S, Grippo AJ, London S, Goossens L, Cacioppo JT. Loneliness: clinical import and interventions. Perspect Psychol Sci. 2015;10:238–49.
Malcolm M, Frost H, Cowie J. Loneliness and social isolation causal association with health-related lifestyle risk in older adults: a systematic review and meta-analysis protocol. Syst Rev. 2019;8:1–8.
Courtin E, Knapp M. Social isolation, loneliness and health in old age: a scoping review. Heal Soc Care Community. 2017;25:799–812.
Steptoe A, Shankar A, Demakakos P, Wardle J. Social isolation, loneliness, and all-cause mortality in older men and women. Proc Natl Acad Sci USA. 2013;110:5797–801.
Stokes AC, Xie W, Lundberg DJ, Glei DA, Weinstein MA. Loneliness, social isolation, and all-cause mortality in the United States. SSM - Ment Heal. 2021;1:100014.
Abdellaoui A, Sanchez-Roige S, Sealock J, Treur JL, Dennis J, Fontanillas P, et al. Phenome-wide investigation of health outcomes associated with genetic predisposition to loneliness. Hum Mol Genet. 2019;28:3853–65.
Elovainio M, Lahti J, Pirinen M, Pulkki-Råback L, Malmberg A, Lipsanen J, et al. Association of social isolation, loneliness and genetic risk with incidence of dementia: UK Biobank Cohort Study. BMJ Open. 2022;12:6–8.
Rødevand L, Bahrami S, Frei O, Lin A, Gani O, Shadrin A, et al. Polygenic overlap and shared genetic loci between loneliness, severe mental disorders, and cardiovascular disease risk factors suggest shared molecular mechanisms. Transl Psychiatry. 2021;11:3.
Hutten E, Jongen EMM, Hajema KJ, Ruiter RAC, Hamers F, Bos AER. Risk factors of loneliness across the life span. J Soc Pers Relat. 2022;39:1482–507.
Boomsma DI, Willemsen G, Dolan CV, Hawkley LC, Cacioppo JT. Genetic and environmental contributions to loneliness in adults: The Netherlands Twin Register study. Behav Genet. 2005;35:745–52.
Day FR, Ong KK, Perry JRB. Elucidating the genetic basis of social interaction and isolation. Nat Commun. 2018;9:1–6.
McLachlan AD. Protein structure and function. Annu Rev Phys Chem. 1972;23:165–92.
Robustelli BL, Newberry RE, Whisman MA, Mittal VA. Social relationships in young adults at ultra high risk for psychosis. Psychiatry Res. 2017;247:345–51.
Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious orders study and rush memory and aging project. J Alzheimers Dis. 2018;64:S161–89.
Wingo AP, Liu Y, Gerasimov ES, Gockley J, Logsdon BA, Duong DM, et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nat Genet. 2021;53:143–6.
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53.
Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science (80-). 2018;362:eaat8464.
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29.
Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8.
Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–52.
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018;7:1–29.
Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, et al. Mendelian randomization. Nat Rev Methods Prim. 2022;2:6.
Hemani G, Tilling K, Smith GD. Orienting the causal relationship between imprecisely measured traits using genetic instruments. PLoS Genet. 2017;13:e1007081.
Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383.
Timshel PN, Thompson JJ, Pers TH. Genetic mapping of etiologic brain cell types for obesity. Elife. 2020;9:1–45.
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Chen CH, Ferreira JCB, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014;94:1–34.
Jin S, Cao Q, Yang F, Zhu H, Xu S, Chen Q, et al. Brain ethanol metabolism by astrocytic ALDH2 drives the behavioural effects of ethanol intoxication. Nat Metab. 2021;3:337–51.
Li K, Xu E. The role and the mechanism of γ-aminobutyric acid during central nervous system development. Neurosci Bull. 2008;24:195–200.
Longa Z, Medlockc C, Dzemidzic M, Shine Y-W, Goddard AWUD. Decreased GABA levels in anterior cingulate cortex/medial prefrontal cortex in panic disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2013;44:131–5.
Lydiard RB. The role of GABA in anxiety disorders. J Clin Psychiatry. 2003;64:21–7.
Paine TA, Swedlow N, Swetschinski L. Decreasing GABA function within the medial prefrontal cortex or basolateral amygdala decreases sociability. Behav Brain Res. 2017;317:542–52.
He J, Xia M, Tsang WH, Chow KL, Xia J. ICA1L forms BAR-domain complexes with PICK1 and is crucial for acrosome formation in spermiogenesis. J Cell Sci. 2015;128:3822–36.
Ou YN, Yang YX, Deng YT, Zhang C, Hu H, Wu BS, et al. Identification of novel drug targets for Alzheimer’s disease by integrating genetics and proteomes from brain and blood. Mol Psychiatry. 2021;26:6065–73.
Cullell N, Gallego-Fábrega C, Cárcel-Márquez J, Muiño E, Llucià-Carol L, Lledós M, et al. ICA1L is associated with small vessel disease: a proteome-wide association study in small vessel stroke and intracerebral haemorrhage. Int J Mol Sci. 2022;23:3161.
Zhang C, Qin F, Li X, Du X, Li T. Identification of novel proteins for lacunar stroke by integrating genome-wide association data and human brain proteomes. BMC Med. 2022;20:1–11.
Corbett AH. Post-transcriptional regulation of gene expression and human disease. Curr Opin Cell Biol. 2018;52:96–104.
Xiong Y, Shie FS, Zhang J, Lee CP, Ho YS. The protective role of cellular glutathione peroxidase against trauma-induced mitochondrial dysfunction in the mouse brain. J Stroke Cerebrovasc Dis. 2004;13:129–37.
Wong CHY, Bozinovski S, Hertzog PJ, Hickey MJ, Crack PJ. Absence of glutathione peroxidase-1 exacerbates cerebral ischemia-reperfusion injury by reducing post-ischemic microvascular perfusion. J Neurochem. 2008;107:241–52.
Hwang SN, Lee JS, Seo K, Lee H. Astrocytic regulation of neural circuits underlying behaviors. Cells. 2021;10:1–24.
Gao J, Davis LK, Hart AB, Sanchez-Roige S, Han L, Cacioppo JT, et al. Genome-wide association study of loneliness demonstrates a role for common variation. Neuropsychopharmacology. 2017;42:811–21.
Matthews T, Danese A, Wertz J, Odgers CL, Ambler A, Moffitt TE, et al. Social isolation, loneliness and depression in young adulthood: a behavioural genetic analysis. Soc Psychiatry Psychiatr Epidemiol. 2016;51:339–48.
Andreu-Bernabeu Á, Díaz-Caneja CM, Costas J, De Hoyos L, Stella C, Gurriarán X, et al. Polygenic contribution to the relationship of loneliness and social isolation with schizophrenia. Nat Commun. 2022;13:51.
Michalska Da Rocha B, Rhodes S, Vasilopoulou E, Hutton P. Loneliness in psychosis: a meta-analytical review. Schizophr Bull. 2018;44:114–25.
Eglit GML, Palmer BW, Martin AS, Tu X, Jeste DV. Loneliness in schizophrenia: construct clarification, measurement, and clinical relevance. PLoS One. 2018;13:1–20.
Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet. 2016;387:1561–72.
Acknowledgements
We thank all the researchers who provided the original data publicly.
Funding
This study was supported by 1.3.5 projects for disciplines of excellence, West China Hospital, Sichuan University (Grant No. ZYJC21004), the National Natural Science Foundation of China (Grant No. 81871061), and the Postdoctoral Foundation of West China Hospital (Grant No. 2020HXBH041).
Author information
Authors and Affiliations
Contributions
XJG: conceptualization, formal analysis, methodology, writing—original draft; MD: data curation and methodology; MLY: writing—review & editing; WZ: conceptualization; writing—review & editing; supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gu, X., Dou, M., Yuan, M. et al. Identifying novel proteins underlying loneliness by integrating GWAS summary data with human brain proteomes. Neuropsychopharmacol. 48, 1087–1097 (2023). https://doi.org/10.1038/s41386-023-01536-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41386-023-01536-0
- Springer Nature Switzerland AG