Abstract
Leukemias arise from recurrent clonal mutations in hematopoietic stem/progenitor cells (HSPCs) that cause profound changes in the bone marrow microenvironment (BMM) favoring leukemic stem cell (LSC) growth over normal HSPCs. Understanding the cross talk between preleukemic mutated HSPCs and the BMM is critical to develop novel therapeutic strategies to prevent leukemogenesis. We hypothesize that preleukemic-LSCs (pLSCs) induce BMM changes critical for leukemogenesis. Using our AML-murine model, we performed single-cell RNA-sequencing of preleukemic BMM (pBMM) cells. We found normal HSC (nHSC)-regulating LepR+ mesenchymal stem cells, and endothelial cells were decreased, along with increases in CD55+ fibroblasts and pericytes. Preleukemic CD55+ fibroblasts had higher proliferation rates and decreased collagen expression, suggesting extracellular matrix remodeling during leukemogenesis. Importantly, co-culture assays found preleukemic CD55+ fibroblasts expanded pLSCs significantly over nHSCs. In conclusion, we have identified a distinct pBMM and a novel CD55+ fibroblast population that is expanded in pBMM that promote fitness of pLSCs over nHSCs.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) is a clonal, malignant disease of the blood and bone marrow (BM) that is heterogeneous at the molecular, cytogenetic, and clinical levels [1]. Non-random chromosomal abnormalities (e.g., deletions, translocations) are identified in 50-55% of all AML patients. In contrast, about 45-50% of all AML cases are cytogenetically normal (CN-AML) [2,3,4,5]. While advances have been made towards understanding the biology of AML, the prognosis is still very poor and there is an unmet need to develop novel and effective therapies [4, 5]. Over the years it has become evident that along with genetic mutations within the hematopoietic stem and progenitor cell (HSPC) compartment, alterations in the BM microenvironment (BMM) also play an important role in transformation, therapy resistance and relapse [6]. Thus, to develop effective AML therapies, it is critical to understand how the intrinsic genetic alterations in HSPCs cooperate with the BMM to facilitate AML initiation, persistence, and therapy resistance. While alterations in BMM have been described in full blown leukemia, the BMM remodeling that happens during the preleukemic stage by HSCs with predisposing AML genetic mutations have not been fully examined. Most studies on the BM niche have utilized either transgenic or viral transduction murine models that induce non-physiologic expression of strong oncogenes that can cause aggressive disease by itself such as the viral transduction of HSPCs with MLL(KMT2A)-translocations, or PDX models that lack a preleukemic phase. Thus, investigating the changes that occur in the BMM before overt disease is present may help us to identify potential biomarkers or novel factors that contribute to preleukemic clonal expansion and ultimately to leukemia progression. These mechanisms have been difficult to identify previously since full blown disease results in massive blast infiltration of the BM, or they may no longer be required after the preleukemic stage.
Our lab has previously developed a murine model of CN-AML (MllPTD/WT; Flt3ITD/WT, hereafter referred to as PTD;ITD) that recapitulates the human disease [7]. This model develops a sporadic leukemia with ~100% penetrance (latency 4 to >18 months) with a preleukemic phase consisting of normal white blood cell counts (WBC), decreases in red blood cells (RBCs) and absence of AML blasts, thus allowing investigation of changes that occur before full-blown leukemic transformation. Using this model we performed single cell RNA-sequencing (scRNA-seq) and found decreases in LepR+ mesenchymal stromal cells (MSCs) and VE-cadherin+ (CD144+) endothelial cells (ECs) -specifically in arteriolar and arteriole ECs with no differences in sinusoidal ECs, along with increases in CD55+ fibroblasts. In vitro assays and microscopic analysis revealed increases in the proliferation of preleukemic CD55+ fibroblasts, suggesting active expansion and remodeling of the BMM by the preleukemic HSCs. Additionally, we also report altered mRNA and protein expression of collagen genes by the preleukemic CD55+ fibroblasts compared to wild-type (WT). Finally, we observe that CD55+ fibroblasts can support preleukemic HSPC growth in vitro. Together, these data present the first analysis of the preleukemic BMM that differs from previously reported fully transformed AML BM niche [8]. The preleukemic BM niche had decreases in MSCs and ECs with an increase in an activated CD55+ fibroblast population that preferentially expands in preleukemic HSCs.
Results
Global alterations in preleukemic bone marrow niche populations
To determine a relevant time-point to examine changes in PTD;ITD mice that would indicate BM dysfunction, WBC and RBC counts in peripheral blood were monitored every 2 weeks starting from 8 weeks after birth. Preleukemic stage was determined to be when we first observed consistent abnormalities in the peripheral blood (PB) cells without the presence of leukemic blasts in PB and BM. For this murine model, the first hematology alteration we noticed was a persistent decrease in RBC counts. Therefore, the preleukemic state is defined as the timepoint when RBC counts were continually low (RBC counts of less than 6 million/µL in 3 consecutive bleeds), without an elevation in WBC counts, and absence of blasts in blood and BM (Supplementary Fig. S1A). To profile the BM niche in preleukemic mice, non-hematopoietic BM stromal cells from WT and preleukemic PTD;ITD mice (N = 3) were sorted by flow cytometry using the markers CD45- Gr1- CD11b- B220- CD3- CD19- Ter119- CD71- (Fig. 1A, Supplementary Fig. S1B) as previously described and analyzed by droplet-based single cell RNA-sequencing [8]. Data was pre-filtered based on RNA counts and removal of hematopoietic cells, resulting in a pool of 12,444 cells (WT = 5677 cells, PTD;ITD = 6767 cells) used for further analysis. Median number of genes detected per cell were 1531 for WT, 1394 for PTD;ITD. WT and preleukemic PTD;ITD stromal cells were integrated, and unbiased clustering was performed resulting in 17 clusters (Fig. 1B). Cell types were annotated based on expression of cell-specific genes (Fig. 1C, D, Supplementary Table 1) as previously described [8]. We identified MSCs (clusters 0, 1, 2, 3, 8, expressing Lepr, and Cxcl12), endothelial cells (clusters 4, 14, 18, expressing Cdh5, and Pecam1), fibroblasts (clusters 5,6, 10, 12 expressing S100a4), pericytes (cluster 7 expressing Atp1b2), osteo-lineage cells (OLCs, clusters 9, 13, 16 expressing Bglap), and chondrocytes (cluster 11, expressing Acan). We then analyzed the percentages of cells across clusters and between the samples.
A Schematic representation of experiment. Preleukemic PTD;ITD or age- and sex-matched WT mice were sacrificed. Femurs and tibias were collagenase digested and Lin- CD45- Ter119- CD71- stromal cells were sorted by flow cytometry. Sorted stromal cells were analyzed by 10x scRNA-seq. B t-Distributed stochastic neighbor embedding (t-SNE) of stromal cells, colored by clustering and annotated based on gene signature. C Dot plot of top expressed genes in stromal cells by clusters. D t-SNE of stromal cells by expression of key cell-type marker genes. E t-SNE of stromal cells colored by condition (WT, blue; preleukemic PTD;ITD, red). F Percentage of stromal cells across each cluster in WT and preleukemic PTD;ITD cells.
We found decreases in percentages of LepR+ MSCs (cluster 0) and arterials ECs (cluster 4) with increases in percentages of fibroblasts (clusters 5, 6, 12) and pericytes (cluster 7) in preleukemic BMM compared with their controls (Fig. 1E, F). Furthermore, we analyzed the proliferation status of stromal cell subpopulations using the cell cycle scoring function included in the Seurat package. Overall, we show decreases in proliferation of stromal cells (Supplementary Fig. S2A) with decreases in the percentage of cycling cells (cells in S and G2M phase) in preleukemic LepR+ MSC and EC clusters, with an increase in preleukemic fibroblasts compared to WT (Supplementary Fig. S2B). We confirmed these findings using a validation cohort in which scRNA-seq was performed 3 additional preleukemic PTD;ITD mice and 3 control WT mice (Supplementary Fig. S3). These data demonstrate substantial changes in the preleukemic BMM that are distinct from WT.
LepR+ MSC clusters 0 and 8 are decreased in the BMM of preleukemic mice
LepR+ MSCs are multipotent stromal cells that can differentiate into osteo-lineage cells, adipocytes, and chondrocytes [9]. They also play a crucial role in regulating HSCs by secreting critical niche factors including Cxcl12, and Kit ligand [10]. Several studies have shown diverse roles of LepR+ MSCs in AML, either promoting or inhibiting AML cell proliferation through induction of proliferation or apoptosis respectively [11]. Since they play an important role in hematopoiesis and AML, we analyzed their presence within the preleukemic BM niche.
We identified 5 clusters of LepR+ MSCs - clusters 0, 1, 2, 3, and 8, expressing canonical LepR+ MSC markers [8]: Lepr, Cxcl12, Kitl, (Fig. 2A–C). These cells comprised most of the stromal cells in normal as well as preleukemic BM. Consistent with their role in HSC regulation, all LepR+ MSC clusters expressed HSC niche factors Cxcl12, Ccl2, and Cxcl14, Ccl19 (Fig. 2D). First, we determined whether there were any differences in the percentages of LepR+ MSC clusters in preleukemic PTD;ITD bone marrow compared with control. We found significant decreases in the cell percentages only in subclusters 0 (~50%) and 8 (~40%) LepR+ MSCs in preleukemic PTD;ITD BM compared to control (Fig. 1F). Since we identify decreases in LepR+ MSC clusters 0, and 8 in preleukemic BM, we further defined these LepR+ MSC subpopulations by analyzing the markers expressed by all cells in these clusters. Upon analysis of top markers expressed in LepR+ MSC cluster 0 (Supplementary Fig. S4), we found these cells express Nme2 and Lgals1, which have been previously shown to be important for anti-inflammatory response [12], and S100a6, which is required for HSCs self-renewal and regeneration [13]. Similarly, we defined subcluster 8 and found that all cells in this cluster express osteo-lineage progenitor cell markers Sp7 and Alpl, which ultimately give rise to osteo-lineage cells that play a crucial role in HSC maintenance [8]. Since LepR+ MSC clusters 0 and 8 are decreased in preleukemic PTD;ITD mice, these data suggests that the subset of LepR+ MSCs required for maintenance and self-renewal of normal HSCs is lost within the preleukemic BM niche.
A t-SNE of stromal cells, highlighting LepR+ MSC clusters. B t-SNE of stromal cells by expression of key MSC marker genes, along with the corresponding violin plot distributions of expression levels across clusters. C t-SNE of LepR+ MSC clusters. D Heatmap of top expressed genes (rows) in LepR+ MSC clusters in the cells of each cluster (columns) ordered by gene categories. E Differentially expressed genes in preleukemic PTD;ITD LepR+ clusters compared to WT.
Finally, to understand transcriptomic changes within all LepR+ MSCs in preleukemic niche, we analyzed genes differentially expressed in all LepR+ MSC clusters in preleukemic BM compared to WT. Gene expression analysis revealed increased expression of genes important for cell adhesion- Spon1 (~2-fold increase, P < 0.01), cytokines and growth factors- Ccn1, (~1.5-fold increase, P < 0.01) Cxcl14, (~1.2-fold increase, P < 0.01), and Cxcl1 (~2-fold increase, P < 0.01), and transcription factors- Snai2 (~1.3-fold increase, P < 0.01), Id3 (~1.5-fold increase, P < 0.01), and Jund (~1.5-fold increase, P < 0.01) (Fig. 2E). We also confirmed the above findings in our validation cohort (Supplementary Fig. S5). These data together indicate substantial changes in the cluster cell percentages and gene expression in the preleukemic LepR+ MSCs compared to normal BMM.
Gain in extracellular matrix (ECM)-secreting osteo-lineage cells (OLCs) in the preleukemic niche
MSCs can differentiate into osteo-lineage cells that comprise of osteoprogenitors and mature osteoblasts. Osteoblasts play a critical role in HSC maintenance, and differentiation and have been implicated in many diseases such as multiple myeloma and AML [14, 15]. Studies have reported that the leukemic BM niche is marked by deficiencies in osteogenesis and loss of mineralized bone, that ultimately disrupts the normal endosteal physiology. Given the importance of osteoblasts in hematopoiesis and leukemogenesis, we investigated the osteo-lineage cells in preleukemic BM.
We identified subclusters 9, 13, and 16 as osteo-lineage cells (OLCs), that express the definitive osteo-lineage markers [8]: Runx2, Bglap, and Sp7 (Supplementary Fig. S6A, B), secreted cytokines (Ccl4, Ccl5, Ccl6, and Cxcl10) and growth factors (Bmp3) (Supplementary Fig. S6C). We analyzed whether there was lineage skewing in OLC population percentages in preleukemic PTD;ITD bone marrow compared to WT and found only an increased percentage of cluster 9 in preleukemic PTD;ITD stroma (~1.5-fold increase) (Fig. 1F). To define each cluster and understand the importance of OLC cluster 9 within the preleukemic niche, we identified the top expressed genes in each subcluster of OLCs in WT and preleukemic PTD;ITD BM and categorized them into secreted factors, extracellular matrix (ECM) genes, enzymes, and transcription factors. We found that cluster 9 had increased expression of ECM collagen genes (Col1a1, Col2a1, Col5a2. Col11a1, Col11a2, and Col12a1) compared to other OLC clusters (Supplementary Fig. S6C), indicating that there is a gain in ECM-secreting OLCs in the preleukemic niche. This suggests remodeled ECM within preleukemic niche might be important for leukemogenesis.
Vascular remodeling of preleukemic niches is different than in full-blown leukemias
BM endothelial cells (ECs) make up the vasculature within the BM that forms an essential component of the BMM. While the sinusoidal vessels act as a site for differentiation of certain hematopoietic progenitors, and as a conduit for mature hematopoietic cells to peripheral circulation, the endosteal arteries and arterioles are important for maintaining HSC quiescence [16]. In the BM of AML patients, BM microvascular density increases, which ultimately promotes the proliferation and mobilization of AML blasts into circulation [17]. We wanted to analyze whether vascular remodeling occurred within the preleukemic BM and characterize these changes.
Consistent with previous studies, we identified 3 EC clusters: 4, 14, and 18, based on the expression of established EC marker genes: Cdh5, Pecam1, and Emcn (Fig. 3A). To determine the subtypes of ECs, we analyzed expression of Vwf, Ly6ahigh for arterial (cluster 4), LepR for arteriolar (cluster 14), and Flt4 for sinusoidal (cluster 18) ECs as previously described (Fig. 3B,C, Supplementary Fig. S7) [8]. To determine changes in preleukemic BM vasculature, we first analyzed the percentage of cells in each EC cluster in preleukemic PTD;ITD and WT BM. Contrary to what is observed in the BM of full-blown AML [8], we found a ~2.8 decrease in arterial ECs (cluster 4) and ~2-fold decrease in arteriolar ECs (cluster 14), with no difference in the percentage of sinusoidal ECs (cluster 18) in preleukemic PTD;ITD bone marrow compared to WT (Fig. 1F). Finally, we identified differentially expressed genes in preleukemic PTD;ITD EC clusters compared to WT. We identified chemokine Cxcl9 was upregulated in PTD;ITD ECs compared to WT (~1.5-fold increase, P < 0.01), which has been shown to induce chemotaxis, and disrupt endothelial barrier function previously [18]. Ccn2 (~1.5-fold decrease, P < 0.01), known to regulate endothelial cell adhesion to pericytes [19], and Cacybp (~1.3-fold decrease, P < 0.01), previously shown to inhibit vascular remodeling [20], were decreased in preleukemic PTD;ITD ECs compared to WT ECs (Fig. 3D), confirming vascular remodeling of BM niche in preleukemic PTD;ITD BM. Finally, we confirmed the EC findings in our validation cohort (Supplementary Fig. S8).
A t-SNE of non-hematopoietic cells, and expression of key endothelial cell marker genes, along with the corresponding distributions of expression levels across clusters. B t-SNE of three subclusters in the endothelial cell cluster. C Expression of top expressed genes (rows) in the cells of each cluster (columns) ordered by gene categories. D Differentially expressed genes in preleukemic PTD;ITD EC clusters compared to WT.
Pericytes are increased in preleukemic bone marrow niche
Pericytes are mural cells that are typically found along ECs, and in addition to providing structural support to vasculature also play an important role in regulating hematopoiesis [21]. While pericytes have not been described in detail in AML, preliminary studies have shown a decrease in pericyte coverage of BM vasculature in bone marrow biopsies of AML patients [22]. To determine whether pericytes are altered in the preleukemic niche, we analyzed the pericytes population in our dataset.
We identified cluster 7 as pericytes based on Atp1b2 expression [8] (Fig. 4A, B). Upon analysis of top markers expressed by all pericytes, we found cell adhesion markers Ninj1, enzymes Cdk6, Khk, and Gclm and transcription factors Ikzf1, Gata1, and Hdgf expressed in pericytes from WT and preleukemic BM (Fig. 4C). To determine if the perivascular niche is changed in preleukemic PTD;ITD BM, we first analyzed the percentage of pericytes in preleukemic BM compared to WT. Contrary to loss of pericytes in AML BM niche [23], we observed a ~2.7-fold increase in percentage of pericytes in preleukemic PTD;ITD BM compared to WT BM (Fig. 1F), suggesting a unique perivascular niche during leukemogenesis. Finally, to understand the transcriptomic changes in preleukemic pericytes, we identified the differentially expressed genes in pericytes in PTD;ITD bone marrow compared to WT, and categorized them into transcription factors, enzymes, and transmembrane signal receptors. Transcription factors Fosb (~2.6-fold decrease, P < 0.01), Klf2 (~3.4-fold decrease, P < 0.01), and Klf6 (~2.2-fold decrease, P < 0.01), known to regulate pericyte recruitment [24] were decreased in preleukemic PTD;ITD bone marrow compared to WT (Fig. 4D), confirming an altered perivascular niche in preleukemic BM.
A t-SNE of stromal cells, highlighting pericyte cluster. B t-SNE of stromal cells by expression of key pericyte marker gene, along with the corresponding distributions of expression level across clusters. C Heatmap of average expression of top expressed genes (rows) of pericyte cluster in the cells of each cluster (columns) ordered by gene categories. D Volcano plot of differentially expressed genes in preleukemic PTD;ITD pericyte cluster compared to WT.
Bone marrow CD55+ fibroblasts are increased in preleukemic niche
Bone marrow fibroblasts are stromal cells that provide mechanical support to HSPCs by secreting extracellular matrix and remodeling of extracellular matrix components into a highly structured network, thereby regulating hematopoiesis [25]. Several studies using human bone marrow fibroblasts have shown that they can protect AML blasts from undergoing apoptosis and cause resistance to therapy in vitro [26, 27]. Given their importance in AML pathogenesis and response to therapy, we analyzed the BM fibroblasts in the preleukemic niche.
We identified 4 fibroblast subclusters: 5, 6, 10, and 12, based on the expression of the fibroblast marker genes: S100a4, Sema3c, and Dcn [8] (Fig. 5A, B). To determine the changes in fibroblast populations within the preleukemic BM, we first analyzed population differences compared to WT BM. Interestingly, we found an increase in fibroblast clusters 5 (~4.8-fold increase), 6 (~3.7-fold increase), and 12 (~2.5-fold increase), while a decrease in fibroblast cluster 10 (~2-fold decrease) (Fig. 1F). To better understand the different fibroblast clusters, we decided to characterize them based on gene expression. All cells in fibroblast clusters 6, and 12 expresses Spi1, a gene previously shown to be an essential regulator of pro-fibrotic gene expression program in fibroblasts [28] (Supplementary Fig. S9A). Cluster 10, which is lost in the preleukemic BM, showed elevated expressions of ECM genes which includes: Col3a1, Col6a3, Fbln2, Lgals1, and Tnxb (Fig. 5C). This suggests that ECM secreting fibroblasts modulate the preleukemic ECM. Additionally, upon analyzing the differentially expressed genes in preleukemic PTD;ITD fibroblast clusters, we found decreased expression of multiple collagen genes- Col1a1 (~4.8-fold decrease, P < 0.01), Col1a2 (~3.6-fold decrease, P < 0.01), Col3a1 (~4.3-fold decrease, P < 0.01), Col4a1 (~1.8-fold decrease, P < 0.01), Col5a2 (~2.5-fold decrease, P < 0.01), in preleukemic PTD;ITD fibroblasts compared to WT (Fig. 5D), confirming a loss of ECM in preleukemic niche.
A t-SNE of stromal cells, highlighting fibroblast clusters. B t-SNE of stromal cells by expression of key fibroblast marker genes, along with the corresponding violin plot distributions of expression levels across clusters. C Heatmap of expression of top expressed genes (rows) of fibroblasts clusters in the cells of each cluster (columns) ordered by gene categories. D Volcano plot of differentially expressed genes in preleukemic PTD;ITD fibroblast clusters compared to WT. E Representative immunofluorescence staining for CD45 (green), Ter119 (green), and CD55 (red) in bone sections from WT and preleukemic mice. Nuclei are counterstained with DAPI. Images are representative of N = 3 per group. (Right) Quantification of number of CD55+ cells per field in WT and preleukemic BM.
Since we observed an overall increase in BM fibroblasts within the preleukemic niche, we analyzed expression of cell surface markers in order to investigate them further. We identified Cd44, and Cd55 to be up regulated in preleukemic PTD;ITD cells (Supplementary Fig. S9B). We validated this finding by performing immunofluorescence on femur sections from preleukemic PTD;ITD and WT mice. We observe an increase in CD45- Ter119- CD55+ fibroblasts in bone marrow of preleukemic PTD;ITD mice compared to WT (Fig. 5E). Hence, we identified an expansion of CD55+ fibroblasts within the preleukemic BM.
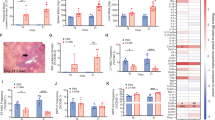
To further characterize the CD55+ fibroblasts and understand their possible role in leukemogenesis, we first sorted CD55+ stromal cells from WT and preleukemic PTD;ITD mice and performed scRNA-seq. Upon clustering, we identified 6 sub-clusters within CD55+ fibroblasts (Supplementary Fig. S10A). Upon analysis of distribution of cells across clusters, we found a loss of cluster 2 and increase in clusters 3, 4, and 5 in PTD;ITD CD55+ fibroblasts compared to WT fibroblasts (Supplementary Fig. S10B, C). Further analysis identified that CD55+ fibroblast cluster 2 had decreased proliferation score compared to other clusters, suggesting that there is a loss of non-cycling CD55+ fibroblasts in preleukemic PTD;ITD BM (Supplementary Fig. S10D). We also confirmed the scRNA-seq findings in our validation cohort (Supplementary Fig. S11). To further validate the scRNA-seq findings, we isolated CD55+ fibroblasts from WT and preleukemic PTD;ITD BM and cultured them in vitro. We confirmed that in vitro cultured P2 fibroblasts expressed CD55 by flow cytometry (Supplementary Fig. S9C). Preleukemic CD55+ fibroblasts showed increased growth in vitro compared to WT CD55+ fibroblasts as analyzed by MTS assay (Fig. 6A). Furthermore, upon analysis of proliferation marker Ki67 by immunostaining CD55+ fibroblasts in BM, we observed an increased percentage of Ki67 + CD55+ fibroblasts in preleukemic BM (Fig. 6B) confirming increased proliferation of CD55+ fibroblasts in the preleukemic niche. These results support the increased percentage of CD55+ fibroblasts identified in the scRNA-seq data. We next validated the changes in extracellular matrix (ECM) proteins in preleukemic BM by performing immunofluorescence on femur sections. Consistent with the scRNA-seq data, we observed a decrease in collagen I (Fig. 6C) and collagen IV (Fig. 6D) staining in preleukemic BM compared to WT BM. These data were also validated by immunophenotyping using flow cytometry on PTD;ITD stromal cells compared to WT (Supplementary Figs. S12, 13). Together, these results suggest that the preleukemic BM undergoes ECM remodeling, which may support expansion of LSCs.
A Growth analysis by MTS assay of P2 CD55+ fibroblasts from WT and preleukemic PTD;ITD mice at 24, 48, and 72 h. N = 4 per group. B Representative immunofluorescence staining for Ki67 (green), and CD55 (red) in bone sections from WT and preleukemic PTD;ITD mice. Nuclei are counterstained with DAPI. Images are representative of N = 5 mice per group. (Right, top) Quantification of number of CD55+ cells per field in WT and preleukemic BM. (Right, bottom) Quantification of percentage of Ki67 + CD55+ cells per field in WT and preleukemic BM. C Representative immunofluorescence staining for Collagen I (green), and CD55 (red) in bone sections from WT and preleukemic PTD;ITD mice. Nuclei are counterstained with DAPI. Images are representative of N = 5 mice per group. (Right, top) Quantification of number of CD55+ cells per field in WT and preleukemic BM. (Right, bottom) Quantification of mean fluorescence intensity (MFI) of Collagen I staining per field in WT and preleukemic BM. D Representative immunofluorescence staining for Collagen IV (green), and CD55 (red) in bone sections from WT and preleukemic PTD;ITD mice. Nuclei are counterstained with DAPI. Images are representative of N = 5 mice per group. (Right, top) Quantification of number of CD55+ cells per field in WT and preleukemic BM. (right, bottom) Quantification of mean fluorescence intensity (MFI) of Collagen IV staining per field in WT and preleukemic BM. E Co-culture followed by CFU-C assay. Briefly, cKit+ cells from WT or preleukemic PTD;ITD mice were co-cultured with P2 WT MSCs or P2 CD55+ fibroblasts from WT or preleukemic PTD;ITD mice for 48 h followed by CFU-C assay. Colonies were counted 14 days later. Quantification of total number of colonies (left). Quantification of number of CFU-GEMM colonies (right). N = 6 per group.
To determine whether these observations are seen in other primary mouse models of preleukemia, we analyzed preleukemic BM of previously published AML mouse models- Mx1-Cre; Stag2 knockout (Stag2 KO) mouse model [29], and Cre+Tet2−/−; NrasG12D/wt (Tet2−/−; NrasG12D/wt) mouse model [30] before full-blown disease. Consistent with our findings in preleukemic PTD;ITD BM, Stag2 KO mice, had increases in CD55+ fibroblasts compared to controls, evaluated using flow cytometry and immunofluorescence (Supplementary Fig. S14A–C). Similarly, we see an increase in CD55+ fibroblasts in BM of preleukemic Tet2−/−; NrasG12D/wt mice compared to controls by flow cytometry and immunofluorescence (Supplementary Fig. S15A–C). These data confirm that our observations are not limited to our PTD;ITD mouse model.
To investigate the effect of CD55+ fibroblasts on LSCs, we performed co-culture assays followed by colony-forming unit cell (CFU-C) assay. cKit cells from WT or preleukemic PTD;ITD mice were co-cultured with CD55+ fibroblasts from WT or preleukemic mice, followed by CFU-C assay. We observed an increase in CFU-C colonies when cKit+ cells from PTD;ITD mice were co-cultured with CD55+ fibroblasts from PTD;ITD mice compared to CD55+ fibroblasts from WT mice (Fig. 6E, Supplementary Fig. S16). To determine whether preleukemic CD55+ fibroblasts secrete cytokines that promote the expansion of LSCs, we analyzed cytokines in culture supernatants from preleukemic PTD;ITD CD55+ fibroblasts compared to control. Interestingly, we saw an increase in secretion of inflammatory cytokines IL-6, and RANTES by preleukemic PTD;ITD CD55+ fibroblasts compared to control (Supplementary Fig. S17). Together, these results suggest that CD55+ fibroblasts expand within the preleukemic BM and support expansion of LSCs, thereby enabling leukemic progression.
Finally, to determine the presence of CD55+ fibroblasts after leukemic transformation, we analyzed CD55+ fibroblasts within BM of leukemic PTD;ITD mice. Interestingly, we see a significant decrease in CD55+ fibroblasts within BM of leukemic PTD;ITD mice compared to control mice by flow cytometry and immunofluorescence (Supplementary Fig. S18A, B). These data suggest that CD55+ fibroblast expansion is specific for the preleukemic phase during leukemogenesis.
Discussion
The BM niche has been well characterized under homeostatic conditions and after the onset of leukemia [8]. However, the changes that take place in the BM niche prior to leukemic transformation has not been well studied. This is the first study to extensively characterize the BM niche cellular heterogeneity prior to leukemic transformation, thus allowing us to identify populations that might possibly be involved in promoting leukemic transformation. The PTD;ITD mouse model with a single copy of each mutant allele, has a prolonged latency period, which allows us to investigate changes that occur in the BM niche prior to leukemic transformation. Furthermore, this mouse model harbors both mutations under the control of their respective normal endogenous promoter thereby allowing physiological and temporal expression of mutant alleles. Using this model, we identified 17 stromal cell clusters with differential frequencies and gene expression patterns in the preleukemic BM niche compared to WT.
RBC counts and conditions of anemia have been frequently associated with leukemic transformation. Presence of moderate or severe anemia was identified as a predictor of leukemic transformation in patients with primary myelofibrosis [31]. Furthermore, previous study has shown that a decrease in erythroblasts and mature RBCs precedes leukemic transformation in a xenograft mouse model [32]. Hence to analyze the changes that take place in BM niche prior to leukemic transformation, we performed scRNA-seq on BM stromal cells at the time point when PTD;ITD mice had decreased peripheral blood RBC counts, without an increase in peripheral blood WBC counts.
The BM niche has been characterized in AML patients and in mouse models of AML after the establishment of the disease. Particularly, the anatomical and functional changes in leukemic BM vasculature have been previously characterized. Using PDX model of AML, a significant loss of ECs associated with sinusoids (CD31+Sca1low) as well as an increased number of ECs associated with arterioles (CD31+Sca1high) is observed in leukemic BM [23]. In our study, while we observe changes in gene expression patterns in EC clusters, we identify no observable differences in percentage of BM sinusoidal ECs. We see a decrease in the percentage of arterial and arteriolar ECs, a subset of ECs known to secrete higher levels of niche factors supporting normal HSCs. Our data shows a loss of EC populations known to support normal HSCs in the preleukemic niche, suggesting an alteration of the BM vascular niche to support preleukemic HSPCs instead. Our results suggest that changes in BM EC numbers occur because of leukemic transformation and might not be required for leukemia initiation.
While frequencies of the majority of BM stromal cell populations remained unchanged in our data, we observed striking increases in pericyte and fibroblast populations in preleukemic BM compared to WT. BM fibroblasts have been characterized to some extent in patients with myelofibrosis, however their role in normal and malignant hematopoiesis has not been thoroughly examined. Fibroblasts share markers with MSCs and other BM stromal cells, and lack definitive markers, thus posing a challenge in their characterization. Previously, using scRNA-seq, defined fibroblast clusters have been identified in normal adult BM [8]. We have similarly identified 4 fibroblast clusters in WT and preleukemic BM. We observed a drastic increase in fibroblast percentages in preleukemic BM compared to WT, as validated by imaging. Interestingly, multiple collagen genes were significantly downregulated in preleukemic BM fibroblasts compared to WT, suggesting that the BM extracellular matrix is remodeled by these fibroblasts during the preleukemic stage.
To identify and further characterize the fibroblast population in preleukemic BM, we analyzed the top expressed genes and identified CD55 to be highly expressed in these cells, which was validated by fluorescence microscopy. CD55 or Decay Accelerating Factor (DAF) is a glycosylphosphatidylinositol (GPI)-anchored membrane protein that has been extensively studied as an inhibitor of complement activation [33]. CD55 has been shown to play an important role in the survival and bone-resorption activity of osteoclasts through regulation of Rac activity [34]. However, the role of CD55 and CD55+ fibroblasts in BM remains to be elucidated. This is the first study to report that CD55+ fibroblasts are increased in preleukemic BM and exhibit increased proliferation in vitro and in vivo. Gene expression analysis and microscopy reveal decreased expression of collagen genes, suggesting a remodeling of the ECM in preleukemic BM. Finally, co-culture assays suggest that CD55+ fibroblasts within the preleukemic niche promote expansion of LSCs. However, future studies to fully characterize the exact role of these CD55+ BM in fibroblasts in leukemic progression are still needed.
Methods
Mouse models
PTD;ITD mouse model
Previously developed MllPTD/WT; Flt3ITD/WT mouse model (PTD;ITD) is a knock-in mouse model [7] and age and sex-matched WT controls were used. Briefly, PTD;ITD mice have a single allele of the Mll-PTD mutation under the regulation of the endogenous promoter, while the other allele is WT. Similarly, the mouse model has a single allele of the Flt3-ITD mutation under the regulation of the endogenous promoter. To define preleukemic stage, WBC and RBC counts in peripheral blood were monitored every 2 weeks starting from 8 weeks after birth. The preleukemic stage was defined when RBC counts were consistently low (RBC counts of less than 6 million/uL in 3 consecutive bleeds), without an elevation in WBC counts and absence of blasts in blood and BM. All mice used for the experiments were within ages 20 weeks to 50 weeks old, both male and female. All controls were age and sex-matched to the preleukemic PTD;ITD mice.
Stag2 KO mouse model
Previously developed Mx1-Cre;Stag2Δ/Δ (Stag2 KO) mice [29] and age and sex matched Mx1-Cre controls were used. Briefly, 8–12 weeks old mice (Mx1-Cre;Stag2fl/fl and Mx1-Cre;Stag2wt/wt) were given 4 IP injections of 200uL pIpC (1 mg/mL) every two days and mice were sacrificed 6 weeks after final injection.
Tet2−/− NrasG12D/wt mouse model
Previously developed Cre+ Tet2−/− NrasG12D/wt mice [30] (Tet2−/− NrasG12D/wt) and age and sex matched WT controls were used. Briefly, BM from 8-12 weeks old mice was transplanted into lethally irradiated BoyJ mice. 2 weeks post-transplant, mice were treated with tamoxifen and mice were sacrificed 6 weeks post tamoxifen treatment.
Single cell sequencing and data processing
Single-cell RNA sequencing (scRNAseq) was performed using 10X Genomics 3’ gene expression kit (v3.1, dual indexed). Stromal cells were sorted as previously described [8]. Briefly, WT and preleukemic (absence of full-blown disease) mice (3 mice per group) were sacrificed, and femurs and tibias were cleaned. Bones were pooled and crushed in collagenase (Worthington Chemicals, 0.5 mg/mL) and incubated at 37 °C for 30 min. Cells were strained using a 70 µm filter and RBCs were lysed using ACK lysis buffer on ice for 5 min. Cells were stained with CD45 (Biolegend, Cat#103104), CD11b (Biolegend, Cat#101204), Gr-1 (Biolegend, Cat#108404), B220 (Biolegend, Cat#103204), CD19 (Biolegend, Cat#152408), Ter119 (Biolegend, Cat#116204), Streptavidin-PE (Biolegend, Cat#405203), CD71 (Biolegend, Cat#113820) and DAPI, and stromal cells were sorted by flow cytometry. For the validation cohort, 3 mice per group were sacrificed and bones from individual mice were crushed and processed as mentioned above. Single cell suspensions of sorted stromal cells were loaded into a 10x Genomics Chromium device for microfluidic-based partitioning and 10,000 single cells were targeted for capture. The manufacturer’s instructions were followed for reverse transcription, cDNA amplification, and library preparation according to the v.3.1 3’-single-cell RNA-seq protocol. Sequencing was performed on an Illumina NovaSeq 6000 instrument to generate paired-end sequencing reads, and FASTQ files were generated using Illumina mkfastq v7.0.0 software. 10x Genomics CellRanger software suite v7.0.0 was used following the default parameters, to perform data pre-processing including alignment, filtering, barcode counting and UMI counting. Downstream analyses were performed using Seurat V3 for R [35]. For each dataset, the lowest 5% of cells based on RNA counts were discarded. Then, UMI counts were log-normalized with the NormalizeData Seurat function. For dataset integration, integration anchors were done using the FindIntegrationAnchors function. Standardized (i.e., centered and reduced) expression values were obtained with the ScaleData function of Seurat.
Dimension reduction and clustering
Dimensionality reduction was performed by running a principal component analysis (PCA) on the scaled data using RunPCA Seurat function. The distance matrix was organized into a K-nearest neighbor graph (KNN), partitioned into clusters using Louvain algorithm using the FindClusters Seurat function, and clusters were visualized on a Uniform Manifold Approximation and Projection (UMAP) using the FindNeighbors Seurat function.
Cluster annotation and top expressed genes
Top differentially expressed genes for each cluster were computed using the FindAllMarkers Seurat function, and cell types were annotated based on expression of known markers, as previously described [36]. Top 10 expressed genes were determined in each cluster and heatmap was generated using heatmap function.
Differential gene expression analysis
Top differentially expressed genes in PTD;ITD stromal cells for each cluster were computed using the FindAllMarkers Seurat function. Fold changes in up- and down-regulated genes were plotted using the R package EnhancedVolcano.
Cell cycle analysis
Cell cycle states of single cells in each cluster were identified based on expression of G2/M and S phase markers using the CellCycleScoring function in Seurat package. Percentage of cells in G1, S, and G2M cell cycle states were computed in each cluster of each group.
Immunofluorescence
Femurs and tibias were harvested from euthanized mice and fixed in formalin for 72 h. Bones were decalcified in EDTA solution for 72 h. Bones were paraffin embedded and sectioned. Tumor sections were deparaffinized, sequentially hydrated and antigen retravel was performed using citrate buffer (pH 6.0). Sections were blocked and stained with primary antibodies anti-CD55 (Thermo Scientific, Cat# PA5-89530, RRID:AB_2805632), anti-CD45 (Thermo Scientific, Cat# 14-0451-82, RRID:AB_467251), anti-Ter119 (Thermo Scientific, Cat# 14-5921-82, RRID:AB_467727), anti-Ki67 (Thermo Scientific, Cat#14-5698-82, RRID:AB_2861195), anti-Collagen I (Thermo Scientific, Cat#MA1-26771, RRID:AB_2081889), and anti-Collagen IV (Thermo Scientific, Cat#SAB4200500, RRID:AB_2574486) and secondary antibodies conjugated with Alexa fluor 488, and Alexa Fluor 647. Nuclei were counterstained with DAPI. Images were obtained using the Olympus FV3000 confocal microscope and analyzed using NIH ImageJ software (RRID:SCR_003070).
Cell culture and MTS assay
WT and preleukemic mice (3 mice per group) were sacrificed, and femurs and tibias were cleaned. Bones were crushed in collagenase (Worthington Chemicals, 0.5 mg/mL) and incubated at 37 °C for 30 min. Cells were strained using a 70 µm filter and RBCs were lysed using ACK lysis buffer on ice for 5 min. Cells were stained with biotinylated lineage cocktail of CD45, CD11b, Gr-1, B220, CD19, Ter119, and CD71 for 30 min on ice. Cells were washed and stained with Dynabeads biotin binder (Thermo, Cat# 11047) and incubated on shaker at 4 °C for 30 min. Cells were washed and lineage negative cells were selected using a magnetic stand. Lineage negative cells were stained with biotinylated CD55 antibody (Thermo, Cat#MA5-17676) and incubated on a shaker at 4 °C for 40 min. Cells were washed and stained with Streptavidin-conjugated MACS beads (MACS, Cat#130-090-485) at 4 °C for 15 min followed by positive selection through MACS LS column. CD55+ cells were cultured in RPMI supplemented with 20% FBS, and antibiotics. Media was changed every 3-4 days and cells were passaged every 7–10 days. P2 CD55+ cells were plated in a 96 well plate and MTS assay was performed according to manufacturer’s instructions at 24, 48, and 72 h post plating. For co-culture assay, cKit+ cells were isolated from WT and preleukemic PTD;ITD mice BM using MACS cKit isolation kit according to manufacturer’s protocol, and co-cultured with CD55+ cells at 1:1 ratio from WT or preleukemic PTD;ITD mice for 48 h, followed by colony formation assay as previously described [37]. Briefly, co-cultured cells were harvested, and equal number of cells were seeded into MethoCult (Stem Cell Technologies, GF M3434). Cells were mixed by vortexing and cells in MethoCult was plated into 3 wells of a 6 well plate. Plates were incubated at 37°C and colonies were counted 14 days later.
Statistical analysis
Data analysis on the examined groups was performed using Graph Pad Prism 8.0 software and comparative analysis using a T-test. Data are reported as mean values ± standard error. Statistical significance is as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
Data availability
Data files will be made available upon reasonable request to the corresponding authors.
References
Short NJ, Ravandi F. Acute Myeloid Leukemia: Past, Present, and Prospects for the Future. Clin Lymphoma Myeloma Leuk. 2016;16S:S25–S9.
Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47.
Dohner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136–52.
Marcucci G, Mrozek K, Bloomfield CD. Molecular heterogeneity and prognostic biomarkers in adults with acute myeloid leukemia and normal cytogenetics. Curr Opin Hematol. 2005;12:68–75.
Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol. 2014;32:548–56.
Behrmann L, Wellbrock J, Fiedler W. The bone marrow stromal niche: a therapeutic target of hematological myeloid malignancies. Expert Opin Ther Targets. 2020;24:451–62.
Zorko NA, Bernot KM, Whitman SP, Siebenaler RF, Ahmed EH, Marcucci GG, et al. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood. 2012;120:1130–6.
Baryawno N, Przybylski D, Kowalczyk MS, Kfoury Y, Severe N, Gustafsson K, et al. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell. 2019;177:1915–32.e16.
Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–68.
Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–34.
Lee MW, Ryu S, Kim DS, Lee JW, Sung KW, Koo HH, et al. Mesenchymal stem cells in suppression or progression of hematologic malignancy: current status and challenges. Leukemia. 2019;33:597–611.
Sundblad V, Morosi LG, Geffner JR, Rabinovich GA. Galectin-1: A Jack-of-All-Trades in the Resolution of Acute and Chronic Inflammation. J Immunol. 2017;199:3721–30.
Grahn THM, Niroula A, Végvári Á, Oburoglu L, Pertesi M, Warsi S, et al. S100A6 is a critical regulator of hematopoietic stem cells. Leukemia 2020;34:3323–37.
Edwards CM, Zhuang J, Mundy GR. The pathogenesis of the bone disease of multiple myeloma. Bone. 2008;42:1007–13.
Frisch BJ, Ashton JM, Xing L, Becker MW, Jordan CT, Calvi LM. Functional inhibition of osteoblastic cells in an in vivo mouse model of myeloid leukemia. Blood. 2012;119:540–50.
Kopp HG, Avecilla ST, Hooper AT, Rafii S. The bone marrow vascular niche: home of HSC differentiation and mobilization. Physiology. 2005;20:349–56.
Kuzu I, Beksac M, Arat M, Celebi H, Elhan AH, Erekul S. Bone marrow microvessel density (MVD) in adult acute myeloid leukemia (AML): therapy induced changes and effects on survival. Leuk Lymphoma. 2004;45:1185–90.
Amatschek S, Lucas R, Eger A, Pflueger M, Hundsberger H, Knoll C, et al. CXCL9 induces chemotaxis, chemorepulsion and endothelial barrier disruption through CXCR3-mediated activation of melanoma cells. Br J Cancer. 2011;104:469–79.
Hall-Glenn F, De Young RA, Huang BL, van Handel B, Hofmann JJ, Chen TT, et al. CCN2/connective tissue growth factor is essential for pericyte adhesion and endothelial basement membrane formation during angiogenesis. PLoS One. 2012;7:e30562.
Zhou J, Li F, Yang Y. Protective effects of calcyclin-binding protein against pulmonary vascular remodeling in flow-associated pulmonary arterial hypertension. Respir Res. 2022;23:223.
Mangialardi G, Cordaro A, Madeddu P. The bone marrow pericyte: an orchestrator of vascular niche. Regen Med. 2016;11:883–95.
Weidenaar AC, ter Elst A, Koopmans-Klein G, Rosati S, den Dunnen WF, Meeuwsen-de Boer T, et al. High acute myeloid leukemia derived VEGFA levels are associated with a specific vascular morphology in the leukemic bone marrow. Cell Oncol. 2011;34:289–96.
Passaro D, Di Tullio A, Abarrategi A, Rouault-Pierre K, Foster K, Ariza-McNaughton L, et al. Increased Vascular Permeability in the Bone Marrow Microenvironment Contributes to Disease Progression and Drug Response in Acute Myeloid Leukemia. Cancer Cell. 2017;32:324–41.e6.
Demolli S, Doddaballapur A, Devraj K, Stark K, Manavski Y, Eckart A, et al. Shear stress-regulated miR-27b controls pericyte recruitment by repressing SEMA6A and SEMA6D. Cardiovasc Res. 2017;113:681–91.
Miari KE, Williams MTS. Stromal bone marrow fibroblasts and mesenchymal stem cells support acute myeloid leukaemia cells and promote therapy resistance. Br J Pharmacol. 2024;181:216–37.
Mei S, Zhang Y, Yu L, Chen G, Zi F. Expression and role of fibroblast activation protein α in acute myeloid leukemia. Oncol Rep. 2021;45:641–51.
Jacamo R, Chen Y, Wang Z, Ma W, Zhang M, Spaeth EL, et al. Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of NF-κB mediates chemoresistance. Blood. 2014;123:2691–702.
Wohlfahrt T, Rauber S, Uebe S, Luber M, Soare A, Ekici A, et al. PU.1 controls fibroblast polarization and tissue fibrosis. Nature. 2019;566:344–9.
Viny AD, Bowman RL, Liu Y, Lavallée VP, Eisman SE, Xiao W, et al. Cohesin Members Stag1 and Stag2 Display Distinct Roles in Chromatin Accessibility and Topological Control of HSC Self-Renewal and Differentiation. Cell Stem Cell. 2019;25:682–96.e8.
Kunimoto H, Meydan C, Nazir A, Whitfield J, Shank K, Rapaport F, et al. Cooperative Epigenetic Remodeling by TET2 Loss and NRAS Mutation Drives Myeloid Transformation and MEK Inhibitor Sensitivity. Cancer Cell. 2018;33:44–59.e8.
Vallapureddy RR, Mudireddy M, Penna D, Lasho TL, Finke CM, Hanson CA, et al. Leukemic transformation among 1306 patients with primary myelofibrosis: risk factors and development of a predictive model. Blood Cancer J. 2019;9:12.
Zhang TY, Dutta R, Benard B, Zhao F, Yin R, Majeti R. IL-6 blockade reverses bone marrow failure induced by human acute myeloid leukemia. Sci Transl Med. 2020;12:eaax5104.
Lublin DM, Atkinson JP. Decay-accelerating factor: biochemistry, molecular biology, and function. Annu Rev Immunol. 1989;7:35–58.
Shin B, Won H, Adams DJ, Lee SK. CD55 Regulates Bone Mass in Mice by Modulating RANKL-Mediated Rac Signaling and Osteoclast Function. J Bone Min Res. 2020;35:130–42.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–902.e21.
Giladi A, Paul F, Herzog Y, Lubling Y, Weiner A, Yofe I, et al. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat Cell Biol. 2018;20:836–46.
Bill M, Goda C, Pepe F, Ozer HG, McNeil B, Zhang X, et al. Targeting BRD4 in acute myeloid leukemia with partial tandem duplication of the. Haematologica. 2021;106:2527–32.
Author information
Authors and Affiliations
Contributions
CG, RK, KEM, and AMD designed the study. CG, RK, YB, WL, AR, OB, SD, APU, MK, YA, EK, SK, NS, and AW performed data collection. CG, RK EARG, KEM, and AMD performed data analysis and interpretation. CG, RG, KEM and AMD wrote the manuscript. BM, RLB, ADV, LAM provided mice for experiments. BM, LS, BZ, GM, ERM, RG, RLB, ADV, LAM, KEM, and AMD critically reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All mouse experiments were conducted in compliance with the Institutional Animal Care and Use Committee (IACUC) at The Ohio State University and Huntsman Cancer Institute at the University of Utah.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Goda, C., Kulkarni, R., Bustos, Y. et al. Cellular taxonomy of the preleukemic bone marrow niche of acute myeloid leukemia. Leukemia (2024). https://doi.org/10.1038/s41375-024-02415-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41375-024-02415-3
- Springer Nature Limited