Abstract
Ribosome display is a cell-free technology for the in vitro selection and evolution of proteins encoded by DNA libraries, in which individual nascent proteins (phenotypes) are linked physically to their corresponding mRNA (genotypes) in stable protein-ribosome-mRNA (PRM) complexes1,2 (Fig. 1). Formation of the complexes can be achieved through deletion of the stop codon of the mRNA, stalling the ribosome at the end of translation; the nascent protein is extended by a spacer such as the immunoglobulin Cκ domain or others to allow exit through the ribosome tunnel3. Through affinity for a ligand, the protein-mRNA coupling permits simultaneous isolation of a functional nascent protein and its translated mRNA; the latter is then converted into cDNA by reverse transcription and amplified for further manipulation, repeated cycles or soluble protein expression. Through the use of PCR-generated libraries, avoiding the need for cloning, ribosome display can be used to both screen very large populations and continuously search for new diversity during subsequent rounds of selection. Additionally, the use of cell-free systems allows the selection of proteins that are toxic or unstable in cells, and proteins with chemical modifications. Ribosome display systems using both prokaryotic and eukaryotic cell extracts have been developed1,2. Examples of the application of eukaryotic systems include the selection and evolution of antibody fragments, DNA binding domains, enzymes, interacting proteins and peptides among others3. Here we describe the step-by-step procedure to perform our previously described eukaryotic ribosome display method, which has the distinctive feature of an in situ reverse transcription–PCR (RT-PCR) procedure for DNA recovery from ribosome-bound mRNA1,4,5,6. We also introduce a recent, previously unpublished improvement to the procedure in which in situ reverse transcription is combined with sensitive single-primer PCR technology.

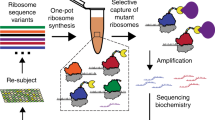
The eukaryotic ribosome display cycle, showing steps of ribosome complex generation from the PCR library in the cell-free expression system, removal of input DNA, selection of complexes by ligand binding, in situ RT-PCR recovery and regeneration of full length cDNA.


Similar content being viewed by others
References
He, M. & Taussig, M.J. ARM complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res. 25, 5132–5134 (1997).
Hanes, J. & Plückthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 94, 4937–4942 (1997).
He, M. & Khan, F. Ribosome display: next-generation of display technologies for production of antibodies in vitro. Expert Rev. Proteomics 2, 421–430 (2005).
He, M. & Taussig, M.J. Ribosome display of antibodies: expression, specificity and recovery in a eukaryotic system. J. Immunol. Methods. 297, 73–82 (2005).
He, M. et al. Selection of a human anti-progesterone antibody fragment from a transgenic mouse library by ARM ribosome display. J. Immunol. Methods 231, 105–117 (1999).
He, M, Cooley, N., Jackson, A. & Taussig, M. Production of human single-chain antibodies by ribosome display. Methods Mol. Biol. 248, 177–189 (2004).
Douthwaite, J.A., Groves, M.A., Dufner, P. & Jermutus, L. An improved method for an efficient and easily accessible eukaryotic ribosome display technology. Protein Eng. Des. Sel. 19, 85–90 (2006).
Lipovsek, D. & Plückthun, A. In vitro protein evolution by ribosome display and mRNA display. J. Immunol. Methods 290, 51–67 (2004).
Zahnd, C., Amstutz, P. & Plückthun, A. Ribosome display selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods 4, 269–279 (2007).
Irving, R. A, Coia, G., Roberts, A., Nuttall, S.D. & Hudson, P.J. Ribosome display and affinity maturation: from antibodies to single V-domains and steps towards cancer therapeutics. J. Immunol. Methods 248, 31–45 (2001).
Rungpragayphan, S. et al. Cloning-independent protein library construction by combining single-molecule DNA amplification with in vitro expression. J. Mol. Biol. 318, 395–405 (2002).
Acknowledgements
Research in the Technology Research Group, Babraham Institute, is supported by BBSRC, Biotechnology and Biological Sciences Research Council UK, and the European Commission Framework 6 Integrated Project MolTools.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
He, M., Taussig, M. Eukaryotic ribosome display with in situ DNA recovery. Nat Methods 4, 281–288 (2007). https://doi.org/10.1038/nmeth1001
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1001
- Springer Nature America, Inc.
This article is cited by
-
Selection and screening strategies in directed evolution to improve protein stability
Bioresources and Bioprocessing (2019)
-
Comparing proteins and nucleic acids for next-generation biomolecular engineering
Nature Reviews Chemistry (2018)
-
Discovery of protein interactions using parallel analysis of translated ORFs (PLATO)
Nature Protocols (2014)
-
Selection of proteins with desired properties from natural proteome libraries using mRNA display
Nature Protocols (2011)