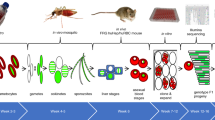
Abstract
Genetic crosses of phenotypically distinct strains of the human malaria parasite Plasmodium falciparum are a powerful tool for identifying genes controlling drug resistance and other key phenotypes. Previous studies relied on the isolation of recombinant parasites from splenectomized chimpanzees, a research avenue that is no longer available. Here we demonstrate that human-liver chimeric mice support recovery of recombinant progeny for the identification of genetic determinants of parasite traits and adaptations.


Similar content being viewed by others
References
Hayton, K. et al. Cell Host Microbe 4, 40–51 (2008).
Ranford-Cartwright, L.C., Hayton, K.L. & Ferdig, M.T. in Malaria Parasites: Comparative Genomics, Evolution and Molecular Biology (eds. Carlton, J.M., Perkins, S.L. & Deitsch, K.W.) Ch. 6, 127–144 (Caister Academic Press, 2013).
Walliker, D. et al. Science 236, 1661–1666 (1987).
Rodhain, J. & Jadin, J. Ann. Soc. Belges Med. Trop. Parasitol. Mycol. 44, 531–535 (1964).
Vaughan, A.M. et al. J. Clin. Invest. 122, 3618–3628 (2012).
Sá, J.M. et al. Proc. Natl. Acad. Sci. USA 106, 18883–18889 (2009).
Sullivan, J.S. et al. Am. J. Trop. Med. Hyg. 69, 593–600 (2003).
Vaughan, A.M. et al. Mol. Biochem. Parasitol. 186, 143–147 (2012).
Trager, W. & Jensen, J.B. Science 193, 673–675 (1976).
Kaushal, D.C., Carter, R., Miller, L.H. & Krishna, G. Nature 286, 490–492 (1980).
Su, X. et al. Science 286, 1351–1353 (1999).
Colwell, R.K. et al. J. Plant Ecol. 5, 3–21 (2012).
Dondorp, A.M. et al. N. Engl. J. Med. 361, 455–467 (2009).
Cheeseman, I.H. et al. Science 336, 79–82 (2012).
Jiang, H. et al. Genome Biol. 12, R33 (2011).
Ariey, F. et al. Nature 505, 50–55 (2014).
Takala-Harrison, S. et al. Proc. Natl. Acad. Sci. USA 110, 240–245 (2013).
Azuma, H. et al. Nat. Biotechnol. 25, 903–910 (2007).
Vaughan, A.M. et al. Cell. Microbiol. 11, 506–520 (2009).
Mita, T. & Jombart, T. Parasitol. Int. 64, 238–243 (2015).
Reilly Ayala, H.B., Wacker, M.A., Siwo, G. & Ferdig, M.T. BMC Genomics 11, 577 (2010).
Oyola, S.O. et al. BMC Genomics 13, 1 (2012).
Quail, M.A. et al. Nat. Methods 9, 10–11 (2012).
Li, H. & Durbin, R. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. Nat. Genet. 43, 491–498 (2011).
Broman, K.W., Wu, H., Sen, S. & Churchill, G.A. Bioinformatics 19, 889–890 (2003).
Acknowledgements
We thank the Center for Infectious Disease Research (formerly Seattle BioMed) insectary and vivarium for mosquito and rodent care, respectively, as well as R. Garcia and M. McDew-White at the Texas Biomedical Research Institute for technical assistance. We thank S. Mikolajczak, E. Wilson and J. Bial for ongoing FRG huHep mouse discussions and M. Macarulay and K. Ushimaru for help with parasite cloning. Thanks also to A. Kaushansky for help with graphics. NIH grants R21 AI 115194–01 to A.M.V. and M.T.F., R37 AI 048071 to T.J.C.A. and Chemistry-Biochemistry-Biology Interface Training Fellowship T32 GM075762 to R.S.P. supported this work, as did Seattle BioMed internal funds to S.H.I.K. The AT&T Genomics Computing Center at Texas Biomedical Research Institute is supported by the AT&T Foundation and the US National Center for Research Resources (NCRR) grant number S10 RR029392, and laboratory work was conducted in facilities constructed with support from Research Facilities Improvement Program grant C06 RR013556 and RR017515 from NCRR.
Author information
Authors and Affiliations
Contributions
A.M.V., S.H.I.K. and M.T.F. conceived, initiated and supervised the project. A.M.V., S.H.I.K., T.J.C.A., R.S.P. and M.T.F. designed experiments and wrote the manuscript. A.M.V., N.C., M.F., L.A.C., R.S.P., I.H.C., S.N. and C.A.H. carried out the experiments. F.H.N. and T.J.C.A. supplied parasites. A.M.V., S.H.I.K., I.H.C., T.J.C.A., R.S.P. and M.T.F. analyzed results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Proof of recombination from the NF54HT-GFP–luc × GB4 cross.
Progeny from the NF54HT-GFP–luc (N) × GB4 (G) cross were cloned after selection with WR99210 (NF54HT-GFP–luc is resistant) and chloroquine (GB4 is resistant). Primers specific for the GFP–luc integration and the chloroquine resistance-associated (CQR) allele pfcrt were used to amplify genomic DNA from ten progeny (1 through 10). Agarose gel electrophoresis shows the presence of both the GFP-luc integration (a) and following ApoI digestion, the presence of the CQR allele, in all progeny (b). Full gels are presented to the right of each panel.
Supplementary Figure 2 Segregation of 7,536 SNPs and microsatellites (MSs) in 14 progeny from the P. falciparum NF54HT-GFP–luc × NHP* experimental genetic cross.
As an example, chromosome 11 is shown in the main body of the text (Fig. 2). Each row represents an individually sequenced chromosome from one of the 14 progeny. Haplotypes inherited from NHP* are shown in black and those from NF54HT-GFP–luc in red, while microsatellite markers from NHP* are shown in yellow and those from NF54HT-GFP–luc in blue. Seventeen of the 22 MS developed for the study are shown; the remaining five were invariant in the two parents. Chromosome regions in white mark the position of recombination breakpoints. Chromosomes are shown from the first to the last genotyped marker, and therefore do not start at zero base pairs. The grey tick marks along the top of each chromosome indicate the position of the segregating SNPs, while the scale (in base pairs) is shown at the base of the figure.
Supplementary Figure 3 Pairwise allele sharing among 14 progeny from the P. falciparum NF54HT-GFP–luc × NHP* experimental genetic cross.
With Mendelian segregation, progeny are expected to share on average 50% of markers that are identical by descent. We calculated the proportion of segregating SNPs at which each of 91 pair-wise combinations of progeny genotypes differ, and plotted the frequency distribution of these values. Segregation was normally distributed and centered on 52% (mean = 52%, s.d. = 8.03, Shapiro-Wilk test, P = 0.27, W = 0.98).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1–4 (PDF 468 kb)
Source data
Rights and permissions
About this article
Cite this article
Vaughan, A., Pinapati, R., Cheeseman, I. et al. Plasmodium falciparum genetic crosses in a humanized mouse model. Nat Methods 12, 631–633 (2015). https://doi.org/10.1038/nmeth.3432
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3432
- Springer Nature America, Inc.
This article is cited by
-
Two transporters enable chloroquine resistance in malaria
Nature Microbiology (2023)
-
Clinically relevant atovaquone-resistant human malaria parasites fail to transmit by mosquito
Nature Communications (2023)
-
Malaria parasites utilize two essential plasma membrane fusogens for gamete fertilization
Cellular and Molecular Life Sciences (2022)
-
The power and promise of genetic mapping from Plasmodium falciparum crosses utilizing human liver-chimeric mice
Communications Biology (2021)
-
Pairwise growth competitions identify relative fitness relationships among artemisinin resistant Plasmodium falciparum field isolates
Malaria Journal (2019)