Abstract
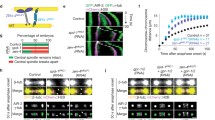
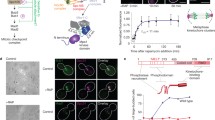
Segregating genetic material along the longest axis of the cell ensures that there is a sufficient distance between daughter chromosomes at the point of cytokinesis. Monitoring the orientation of the mitotic spindle can be subjected to cell cycle controls. In the fission yeast Schizosaccharomyces pombe, the existence of such a cell-cycle checkpoint has been proposed to delay the metaphase to anaphase transition when spindle poles are not properly oriented with respect to the actomyosin ring1. Here we show, by using a fission yeast mutant compromised in its assembly of astral microtubules, that in the absence of astral microtubules short metaphase spindles are unable to orient themselves with respect to the long axis of the cell and are delayed in spindle elongation. This astral defect engages a spindle orientation checkpoint because deletion of the transcription factor Atf1, which is involved in maintaining this checkpoint, allows misaligned asterless metaphase spindles to elongate. We propose that astral microtubules are involved directly in monitoring orientation of the metaphase spindle and in controlling the timing of elongation in fission yeast.




Similar content being viewed by others
References
Gachet, Y., Tournier, S., Millar, J. B. & Hyams, J. S. Nature. 412, 352–355 (2001).
Hagan, I. M. & Hyams, J. S. Cell Motil. Cytoskeleton. 34, 69–75 (1996).
Ding, D. Q., Chikashige, Y., Haraguchi, T. & Hiraoka, Y. J. Cell Sci. 111, 701–712 (1998).
Sparks, C. A., Morphew, M. & McCollum, D. J. Cell Biol. 146, 779–790 (1999).
Decottignies, A. & Nurse, P. J. Cell Sci. 114, 2627–2640 (2001).
Takeda, T. et al. EMBO J. 14, 6193–6208 (1995).
Wilkinson, M. G. et al. Genes Dev. 10, 2289–2301 (1996).
White, J. & Strome, S. Cell. 84, 195–198 (1996).
Carminati, J. L. & Stearns, T. J. Cell Biol. 138, 629–641 (1997).
Kaltschmidt, J. A., Davidson, C. M., Brown, N. H. & Brand, A. H. Nature Cell Biol. 2, 7–12 (2000).
Moreno, S., Klar, A. & Nurse, P. Methods Enzymol. 194, 795–823 (1991).
Balasubramanian, M. K., McCollum, D. & Gould, K. L. Methods Enzymol. 283, 494–506 (1997).
Acknowledgements
We thank Y. Hiraoka, J. Millar, A. Decottignies and K. Gull for materials and reagents; G. Jedd for comments on the manuscript; and members of the fungal laboratories, especially V. Rajagopalan, V. Wachtler, N. Naqvi and K. Wong for discussions. Our research was supported by A*STAR Singapore and Temasek Life Sciences Laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary figure and table
Figure S1 Mia1+ gene from S. pombe has an overall sequence homology to the hypothetical protein NCU01413.1from N. crassa. (PDF 214 kb)
Supplementary table 1
Movie 1
A time-lapse sequence of spindle dynamics in α-tubulin-EGFP expressing wild type S. pombe. (MOV 900 kb)
Movie 2
A time-lapse sequence of spindle dynamics in α-tubulin-EGFP expressing mia1Δ S. pombe cell. (MOV 1291 kb)
Movie 3
A time-lapse sequence of spindle dynamics, visualized using a Sid2p-EGFP expressed in wild type S. pombe. (MOV 672 kb)
Movie 4
A time-lapse sequence of spindle dynamics, visualized using a Sid2p-EGFP expressed in mia1Δ cell. (MOV 1068 kb)
Movie 5
A time-lapse sequence of spindle dynamics in α-tubulin-EGFP-expressing mia1Δatf1Δ cell. (MOV 926 kb)
Rights and permissions
About this article
Cite this article
Oliferenko, S., Balasubramanian, M. Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat Cell Biol 4, 816–820 (2002). https://doi.org/10.1038/ncb861
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb861
- Springer Nature Limited
This article is cited by
-
Microtubules and Alp7–Alp14 (TACC–TOG) reposition chromosomes before meiotic segregation
Nature Cell Biology (2013)
-
Disruption of Tacc3 function leads to in vivo tumor regression
Oncogene (2012)
-
Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome
Nature Communications (2011)
-
Nucleocytoplasmic transport of Alp7/TACC organizes spatiotemporal microtubule formation in fission yeast
EMBO reports (2009)
-
Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast
Nature (2007)