Abstract
Familiar clustering of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) has been frequently reported. However, limited information is available about the underlying molecular mechanisms in HBV-related HCC patients with family history of HCC. In our previous study, Agilent miRNA Base 16.0 microarray showed miRNA profiles of the plasma of HBV-related HCC patients who had a family history of HCC. This study aims to explore the expression, function, and mechanisms of miR-3188 in HCC that might provide novel insights into the role of family history on the risk of HCC. The expression levels of miR-3188 were markedly overexpressed in HCC tissues, HBV transgenic mice, and HepG2.215 cells. We knocked out miR-3188 in HCC cell lines using the CRISPR/Cas9 system, and demonstrated that miR-3188 knockout (KO) suppressed cell growth, migration, and invasion, and inhibited xenografts tumor growth in nude mice. Next, we determined that miR-3188 KO exerts antitumor functions by directly repressing ZHX2. It has been reported that HBV X protein (HBx) plays a critical role in HBV-related HCC, promoting CREB-mediated activation of miR-3188 and activation of Notch signaling through repressing ZHX2. Finally, we verified that ZHX2 functions as a transcriptional repressor to Notch1 via interaction with NF-YA. Our data demonstrate that the HBx–miR-3188–ZHX2-Notch1 signaling pathway plays an important role in the pathogenesis and progression of HBV-related HCC with family history of HCC. These findings have important implications for identifying new therapeutic targets in HBV-related HCC.
Similar content being viewed by others
Main
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third most common cause of cancer-related deaths worldwide.1 Most cases of HCC are associated with hepatitis B virus (HBV) infections in East Asia. It has been reported that the lifetime risk of HBV carriers to develop HCC was much higher than healthy individuals.2, 3 As the prognosis of HCC remains unsatisfactory even after curative resection, further investigation on molecular and hereditary mechanism is needed to achieve better prognosis.4
Familiar clustering of HCC has been frequently documented in East Asia. A recent study has found that family history of HCC multiplies the risk of HCC in patients with HBV infection, and recommends that HBV patients need further refinement of HCC surveillance strategy that could be personalized based on the status of family history.5, 6 Accumulating evidence revealed that genetic factors may contribute to familiar clustering of HCC.7, 8 However, the underlying mechanism of the pathogenesis and progression of HBV-related HCC associated with family history remains to be not clear.
MicroRNAs (miRNAs) are small noncoding RNAs that behave as a ubiquitous feature to modulate key cellular processes involved in carcinogenesis.9 Evaluation of changes in miRNA levels show promise as biomarkers for early detection, diagnosis, and prognosis.10, 11 Increasing evidence also supports the role of miRNAs as potential therapeutic targets for multiple diseases.12 Our previous study detected miRNA profiles of the plasma of HBV-associated HCC patients with a family history of HCC that showed that miR-3188 was the markedly overexpressed miRNA in previous chip profiles and its functions have not been elucidated in HCC.13 Therefore, the key signatures of miR-3188 deserve further investigations.
In this study, we explored the expression pattern of miR-3188 in HCC and its clinical significance, and sought to manipulate miR-3188 expression in human HCC cell lines by the CRISPR/Cas9 genome editing technology. The role of miR-3188 in HCC and the underlying mechanism of miR-3188 regulation and cancer-related signaling pathways were further investigated.
Results
MiR-3188 is overexpressed in HBV-associated HCC and its expression is associated with poor clinical outcome
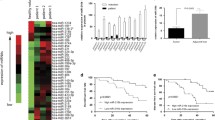
In our previous study, Agilent miRNA Base 16.0 microarray (Agilent Technologies, Santa Clara, CA, USA) showed that miR-3188 was 124.57-fold overexpressed in the plasma of patients with HBV-associated HCC compared with that of healthy volunteers. This result was validated by measuring miR-3188 expression in the plasma of 10 patients with HBV-associated HCC and 10 healthy volunteers using qRT-PCR (P<0.01; Figure 1a). Furthermore, miR-3188 expression was detected using qRT-PCR in 150 pairs of HBV-associated HCC patients’ tumor tissues and corresponding adjacent liver tissues that revealed that miR-3188 expression was significantly overexpressed in HCC tissues than in adjacent liver tissues (P<0.01; Figure 1b).
MiR-3188 is overexpressed in HCC tissues and plasma, and the expression is associated with the prognosis of HCC patients. (a) qRT-PCR analysis of the expression levels of miR-3188 in plasma between 10 pairs of HCC patients and healthy volunteers. The miR-3188 expression levels in healthy volunteer plasma were significantly lower than those in HCC patients’ plasma (5.23±1.45 versus 127.9±18.92, n=10, P<0.01). (b) qRT-PCR analysis of the expression levels of miR-3188 in 150 pairs of HCC patients’ tumor tissues and corresponding adjacent liver tissues. The expression of miR-3188 in tumor tissues was significantly higher than that in adjacent liver tissues. After curative liver resection, the cumulative disease-free survival (c) and overall survival (d) rates in patients with high miR-3188 expression were significantly lower than those with low miR-3188 expression. **P<0.01
Kaplan–Meier log-rank analysis indicated that HBV-associated HCC patients with high miR-3188 expression had significantly shorter median disease-free survival (DFS) (15 versus 39 (months), P<0.01, Figure 1c) and significantly shorter median overall survival (OS) compared with those with low miR-3188 expression (28 versus 66 (months), P<0.01, Figure 1d). Then, we examined the correlation between miR-3188 expression level and the clinicopathological variables in patients with HBV-associated HCC. MiR-3188 overexpression was significantly correlated with elevated serum HBV DNA and α-fetaprotein (AFP) level (P<0.05; Supplementary Table 2). We also analyzed the prognostic factors influencing OS. Univariate analysis showed that overexpression of miR-3188, elevated serum HBV DNA level, elevated serum AFP level, multiple tumors, vascular invasion, and TNM stage were associated with worse OS. Multivariate analysis using Cox’s proportional hazards model showed that overexpression of miR-3188, vascular invasion, and TNM stage were independent prognostic factors for OS (Supplementary Table 3).
MiR-3188 knockout by CRISPR/Cas9
Considering the terminal loop region controls microRNA processing by Drosha and Dicer, single-guide RNA (sgRNA) was designed within Drosha processing sites using CRISPR DESIGN (Supplementary Figure 1a). The sgRNA was subcloned into the plasmid lentiCRISPR v2 containing two expression cassettes, hSpCas9 and the chimeric guide RNA. Then, we transfected the plasmids into HCC cell lines, HepG2 and SMMC7721, and performed T7 endonuclease I assay to detect the CRISPR efficiency 72 h after transfection. Indels generated by CRISPR/Cas9 indicated the cleavage of genome DNA (Supplementary Figure 1b). DNA sequencing confirmed the sequence of indels adjacent to PAM sequence (Supplementary Figure 1c). As shown in Supplementary Figure 1d, in the HepG2 and SMMC7721 cells transfected with designated CRISPR/Cas9 sgRNA compared with the control vectors, the expression levels of mature miR-3188 declined up to 72.4 and 91.1%, and the expression levels of pri-miR-3188 increased up to 83.1 and 91.2%. Knockout (KO) of miR-3188 by CRISPR/cas9 can markedly upregulate the expression of ZHX2 (Supplementary Figure 1e).
MiR-3188 KO suppresses cell growth and colony formation, inhibits cell cycle progression, and triggers apoptosis in HCC cells
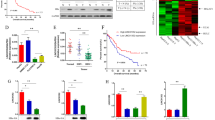
To study the effects of miR-3188 KO on HCC cells, miR-3188 KO stably transfected sgRNA-miR-3188 clones and control clones were used for further experiments. The qRT-PCR analysis confirmed that miR-3188 KO cell clones showed much lower level of miR-3188 than those of control cells after 1 month of transfection (P<0.01, Figure 2a). The CCK-8, colony formation, and anchorage-independent cell growth assays revealed that downregulation of miR-3188 significantly inhibited cell growth and colony formation of HepG2 and SMMC7721 cells (Figures 2b–d).
Antitumor effects of miR-3188 knockout (KO) in HCC cell lines. (a) Relative expression of miR-3188 detected by qRT-PCR in HepG2 and SMMC7721 cell lines stably transfected with CRISPR/Cas9 constructs targeting miR-3188 after 1 month of transfection. (b) By CCK-8 assay, miR-3188 KO suppressed cell viability in HCC cell lines (at 24, 48, and 72 h). (c) The representative images of plate colony formation and anchorage-independent growth in HepG2 cells. (d) Histograms show the colony numbers of the indicated clones. (e) MiR-3188 KO induced G0–G1-phase arrest determined by cell cycle analysis using propidium iodide staining flow cytometry 24 h after transfection. (f) Histograms indicate the percentage of cells in G0–G1, S, and G2–M cell cycle phases. (g) Flow cytometry analysis demonstrated miR-3188 KO induced apoptosis using Annexin V-PE/7AAD staining flow cytometry 48 h after transfection. (h) Histograms indicate the percentage of apoptotic cells in miR-3188 KO and control clones. (i) Cell migration was evaluated by wound healing assay. Representative images were obtained at time points 0 and 72 h. (j) Migration ability was quantified by measuring gap distance. (k) The representative images of cell migration (24 h after transfection) and invasion (48 h after transfection) across a membrane with 8 mm pores with or without Matrigel. (l) Histograms show the number of cells across the membrane. Each experiment was repeated three times. *P<0.05, **P<0.01
To further investigate the mechanism of cell growth inhibition of miR-3188 KO in HCC cells, we did cell cycle distribution and apoptosis analysis using flow cytometry. The results indicated that miR-3188 KO significantly increased the cell percentages in G0/G1 phase and decreased the cell percentages in S phase (Figures 2e and f). Our data also indicated that miR-3188 KO significantly induced apoptosis in HCC cells. The apoptotic rates of control and miR-3188 KO of HepG2 cells were 3.73±0.71 and 16.73±1.35%, respectively, and the apoptotic rates of SMMC7721 control and miR-3188 KO cells were 4.66±0.66 and 18.39±2.42%, respectively (Figures 2g and h). Analysis of potential cell cycle regulators indicated that downregulation of miR-3188 in HepG2 cells decreased Cyclin A and Cyclin E protein levels. Western blotting showed that miR-3188 KO led to a significant increase of apoptosis (Supplementary Figure 2a). These data suggest that miR-3188 KO suppresses cell cycle progression and induces apoptosis, and this may explain the underlying mechanism of miR-3188 KO-induced cell growth inhibition.
MiR-3188 KO decreases migration and invasion of HCC cells
A wound healing assay for cell migration was performed in miR-3188 KO and control clones of HepG2 and SMMC7721. Expectedly, miR-3188 KO significantly decreased migration in both HCC cells (P<0.01; Figures 2i–j). In addition, the transwell chamber migration and invasion assay revealed that miR-3188 KO significantly reduced migration and invasion in both HCC cells (P<0.01; Figures 2k and l). Moreover, we found that miR-3188 KO inactivated the Notch1 pathway in HepG2 cells (Supplementary Figure 2b). Taken together, miR-3188 KO inhibited malignant features in HCC cells.
ZHX2 is a direct target of miR-3188 in HCC cells
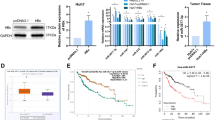
To investigate the miR-3188 target in HCC, we applied miRNA target predicted databases (TargetScan and MiRDB) to identify potential target genes. ZHX2 was identified as a target by both miRNA databases. Schematic diagram of putative has-miR-3188 binding site in the 3′-UTR of ZHX2 is shown in Figure 3a. To determine whether ZHX2 was regulated by miR-3188, qRT-PCR and western blot assays were performed in miR-3188 overexpression or knockout and corresponding control cells. Both mRNA and protein levels of ZHX2 were downregulated by increased miR-3188 or upregulated via miR-3188 KO. (Figure 3b and Supplementary Figure 2b). To further confirm that miR-3188 directly targets ZHX2, we constructed a wild-type ZHX2 3'-UTR luciferase reporter vector. MiR-3188 overexpression significantly reduced the luciferase activity from wild-type ZHX2 3'-UTR luciferase reporter construct. This profound inhibition was abolished when the putative miR-3188 target sequences in the ZHX2 3′-UTR were mutated in the vector, indicating that miR-3188 inhibits ZHX2 by sequence targeting (Figure 3c). Moreover, the abundance of miR-3188 was inversely correlated with low expression levels of ZHX2 in HCC tissues (R2=0.508, P<0.01; Figure 3d). To elucidate whether the reduction of ZHX2 could rescue the antitumor effects of miR-3188 KO, we transfected HepG2 cells with miR-3188 KO plus siZHX2. The reduced ZHX2 expression partially reversed the antitumor effects induced by miR-3188 KO (Figures 3e–l). These results suggested that miR-3188 directly targets ZHX2 expression, and downregulation of ZHX2 can reverse antitumor effects of miR-3188 KO in HCC cells.
MiR-3188 functions as an oncogene in HCC by targeting ZHX2 and activating Notch signaling. (a) Schematic diagram of putative binding sites of miR-3188 on ZHX2 3′-UTR. ZHX2 3′-UTR mutant indicated the 3′- UTR of ZHX2 with the mutation in miR-3188 binding sites. (b) Relative luciferase assay in HepG2 cells showed that miR-3188 significantly suppressed luciferase activity of wild-type reporter constructs, whereas miR-3188 KO significantly promoted luciferase activity of wild-type constructs. (c) MiR-3188 overexpression significantly reduced the luciferase activity from wild-type ZHX2 3'-UTR luciferase reporter construct, whereas this profound inhibition was abolished when the putative miR-3188 target sequences in the ZHX2 3′-UTR were mutated in the vector. (d) Overexpression of miR-3188 was inversely correlated with ZHX2 downregulation (R2=0.508, P<0.01, Pearson’s correlation). (e) When HepG2 cells were co-transfected with miR-3188 KO or control and siZHX2 or siRNA-NC using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction, cell viability of transfected cells was determined by using CCK-8 assay at 24, 48, and 72 h. (f) Cell cycle was detected 24 h after transfection by propidium iodide staining flow cytometry. Bar charts indicated the percentage of cells in G0–G1, S, and G2–M cell cycle phases. (g) Cell apoptosis was determined by Annexin-V/phycoerythrin combined labeling flow cytometry 48 h after transfection. Evaluation of apoptosis was determined by the percentage of apoptotic cell number in total cell number. (h) Migration ability was quantified by measuring gap distance at time points 0 and 72 h. (i) Representative images of plate colony formation and anchorage-independent growth in HepG2 cells are shown. (j) Histograms show the colony numbers of the indicated clones. (k) Representative images of cell migration (24 h after transfection) and invasion (48 h after transfection) across a membrane with 8 mm pores with or without Matrigel are exhibited. (l) Histograms indicate the number of cells across the membrane. All experiments were performed in triplicate. *P<0.05, **P<0.01
MiR-3188 KO inhibits human HCC xenografts growth in nude mice
To examine whether miR-3188 KO could inhibit malignant features of HCC in vivo, subcutaneous tumor models were established in the flanks of athymic nude mice with miR-3188 KO and control clones. We found that miR-3188 KO could markedly inhibit the tumor growth (Figure 4a). The growth curves showed that the average fold increase of miR-3188 KO tumor volumes at the time of killing was much smaller than that in control tumors (HepG2 models: 5.93±0.73 versus 9.79±1.45; SMMC721 models: 9.52±2.63 versus 13±1.6; P<0.01; Figure 4b), and the average weight of miR-3188 KO tumors was significantly lighter than that in control tumors (HepG2 models: 212±21.21 versus 420±24.34; SMMC721 models: 327.4±25.07 versus 528.8±35.86; P<0.01; Figure 4b).
Tumor suppressive effects of miR-3188 KO in vivo. (a) The subcutaneous tumors derived from the miR-3188 KO clones were smaller in size than control clones (n=5 each group). (b) Tumor growth curves show that the tumors derived from the miR-3188 KO clones grew significantly slower than those from control clones at all time points (at 4, 8, 12, 16, 20, 24, and 28 days). Histograms indicate the average tumor weight of each group at the time of killing (n=5 each group). (c) The expression status of ZHX2, Notch signaling, TUNEL, and Ki67 in the xenograft tumor tissues detected by immunohistochemical analysis. (d) The orthotopic tumor implantation assays demonstrated that the liver tumors (right lobe) derived from HepG2-miR-3188 KO clones were markedly smaller in size compared with control group 30 days after orthotopic implantation (n=5 each group). Representative Ki67 staining of miR-3188 KO and control clone generated orthotopic liver tumor tissues. (e) The intraperitoneal tumor implantation assays showed that liver tumors derived from HepG2-miR-3188 KO clones were markedly smaller in size and less in number compared with control clones 21 days after peritoneal injection (n=5 each group). Representative H&E staining of liver tumor tissues derived from miR-3188 KO and control clones. *P<0.05, **P<0.01
Then, we explored the underlying mechanisms of tumor suppression induced by miR-3188 KO. Immunohistochemical staining assay indicated that the Notch-1 signaling pathway was markedly inactivated and expression of Ki67 was reduced, and TUNEL assay demonstrated that apoptotic rate was markedly higher in the resected tumors derived from the miR-3188 KO cells compared with control cells (Figure 4c).
To further evaluate the effect of miR-3188 KO on HCC tumor growth, we established an orthotopic tumor model in nude mice. The results showed that the average tumor volume derived from HepG2-miR-3188 KO cells was significantly smaller than the tumors derived from control cells (2.35±0.26 versus 3.85±0.3; P<0.05; Figure 4d). Immunohistochemical staining study indicated that expression of Ki67 was decreased in the miR-3188 KO tumors compared with the control group.
Moreover, intraperitoneal xenograft models were established to evaluate the effect of miR-3188 KO on HCC tumor growth and metastasis. Compared with the tumors derived from control cells, miR-3188 KO tumors presented the smaller average tumor size (0.43±0.15 versus 1.87±0.31; P<0.01) and less average number (6±2 versus 45.33±7.02; P<0.01; Figure 4e). Consistent with the results above, hematoxylin and eosin staining revealed markedly reduced tumor size and number in tumors derived from miR-3188 KO cells (Figure 4e). Taken together, miR-3188 KO suppressed HCC growth and metastasis in vivo, indicating that the roles of miR-3188 serve as a potential therapeutic target for HCC.
HBx promotes CREB-mediated activation of miR-3188 and downregulates ZHX2
We determined the expression of miR-3188, ZHX2, and HBx by qRT-PCR and western blot assays in HCC tissues, respectively. Our data indicated that HBx and miR-3188 expressions were upregulated and ZHX2 expression was downregulated in 150 HCC tissues, whereas HBV infection was strongly associated with miR-3188 (R2 =0.819, P<0.01) and ZHX2 expression (R2=0.703, P<0.01; Figures 5a and b). Next, we found that the miR-3188 expression levels were significantly higher in liver tissues of 6-month-old HBV transgenic mice than in those of C57/BL6 mice (P<0.01; Figure 5c). The miR-3188 expression was significantly higher in HepG2.215 cells than in HepG2 cells (P<0.01; Figure 5d). Then, we transfected HepG2.215 cells with siHBx, causing the downregulation of miR-3188 expression, and the upregulation of ZHX2 expression (P<0.01; Figure 5d). Furthermore, pcDNA 3.1-HBx was transfected into the HepG2 and SMMC7721 cells, subsequently upregulating miR-3188 expression in a dose-dependent manner (P<0.01; Figure 5e). Taken together, our data suggested that HBx is able to upregulate miR-3188 in HCC.
HBx promotes CREB-mediated activation of miR-3188 and downregulates ZHX2. (a) The results indicated that HBx and miR-3188 expression was upregulated and ZHX2 expression was downregulated in 150 HCC tissues, and HBx was strongly associated with miR-3188 (R2 =0.819, P<0.01) and ZHX2 expression (R2=0.703, P<0.01) (b) Western blot analysis of HBx and ZHX2 expression in four pairs of HCC tumor tissues (T) and their corresponding adjacent nontumor liver tissues (N). (c) The miR-3188 expression of liver tissues were significant higher in HBV transgenic mice than those in C57BL/6 mice (n=3 each group). (d) qRT-PCR analysis for miR-3188 and western blotting analysis for ZHX2 in HepG2 and HepG2.2.15 cells with or without HBx inhibition. (e) qRT-PCR analysis for miR-3188 and western blotting analysis for ZHX2 in HepG2 and SMMC-7721 cell lines transfected with HBx at different doses and controls. (f) HepG2-HBx and SMMC7721-HBx cells were transfected with siCREB or control siRNA and analyzed for miR-3188 expression by qRT-PCR and for ZHX2 expression by western blotting. (g) HepG2 and SMMC7721 cells were transfected with CREB and analyzed for miR-3188 expression by qRT-PCR and for ZHX2 expression by western blotting. (h) Luciferase assay of the putative miR-3188 promoter in HepG2 and SMMC7721 cells transfected with HBx and controls. (i) ChIP analysis showed that HBx promoted CREB occupying on the putative miR-3188 promoter in HepG2 and SMMC7721 cells. All experiments were performed in triplicate. *P<0.05, **P<0.01
It has been reported that HBx is not able to bind directly to DNA, but it is suggested to interact with transcription factors, such as p53 and CREB, to regulate gene expression. Strikingly, we observed that putative CREB-binding sites were located in the miR-3188 upstream via Alggen Promo and JASPAR. Then, we applied siCREB to downregulate CREB in HepG2-HBx and SMMC7721-HBx cells that induced decrease in miR-3188 expression and increase in ZHX2 expression (P<0.01; Figure 5f). In contrast, pcDNA 3.1-CREB was transfected into the cells that triggered increase in miR-3188 expression and decrease in ZHX2 expression (P<0.01; Figure 5g). Furthermore, CREB stimulated the activity of the luciferase reporter gene containing the CREB-binding site, whereas CREB failed to activate the luciferase reporter gene containing mutant CREB-binding site or an upstream site without the CREB-binding site (P<0.01; Figure 5h). To further determine the interaction of HBx with miR-3188 promoter, chromatin immunoprecipitation (ChIP) assay was performed. The results indicated that CREB was recruited to the miR-3188 promoter fragment, and HBx further increased the promoter occupancy of CREB (P<0.01; Figure 5i). In summary, HBx promotes miR-3188 transcription by interacting with CREB.
MiR-3188 KO inactivated the HBx-mediated Notch signaling pathway through restoration of ZHX2
To further explore the mechanism of miR-3188 KO mediated antitumor effects on HCC, we identified potential downstream signaling pathways using western blot assay. We observed that the Notch signaling pathway was inactivated in miR-3188 KO clones and miR-3188 promoted the activation of Notch signaling pathway in HepG2 cells (Supplementary Fig. 2c). In addition, we found that inactivation of Notch signaling pathway induced by miR-3188 KO could be abolished through the inhibition of ZHX2 expression, and activation of Notch signaling pathway via miR-3188 could be suppressed by overexpression of ZHX2 (Supplementary Figure 2c).
It has been reported that Notch signaling was engaged in HBx-related HCC. We found that miR-3188 KO abolished the HBx-mediated activation of the Notch signaling pathway in HepG2 cells, and miR-3188 restored the activity of Notch signaling pathway in HepG2.215 cells treated with siHBx (Supplementary Figure 2d). Furthermore, we transfected siZHX2 into the HepG2 cells, and the Notch signaling pathway was subsequently activated. Then, RO4929097 and γ-secretase inhibitor (DAPT) was used to block siHBx-induced activation of the Notch signaling pathway (Supplementary Figure 2e). Our data suggested that miR-3188 KO abolished the HBx-induced activation of Notch signaling pathway through ZHX2.
ZHX2 functions as transcriptional repressors of Notch1 via interacting with NF-YA
It has been reported that ZHX2 functions as a transcriptional repressor. We transfected BEL7402 cells with pcDNA3.1-ZHX2, causing the downregulation of Notch1 expression in a dose-dependent manner. Notch1 expression increased dose dependently when siZHX2 was transfected into HepG2 cells (P<0.01; Figure 6a). It has been suggested that ZHX2 regulates Notch1 expression. To examine whether ZHX2 regulates Notch1 promoter activity, ZHX2 and wild-type promoter pGL3-Notch1 were co-transfected into BEL7402 cells, whereas siZHX2 and pGL3-Notch1 were co-transfected into HepG2 cells. Our data showed that ZHX2 reduced the relative luciferase activity of Notch1 dose dependently, whereas siZHX2 increased the relative luciferase activity of Notch1 dose dependently (P<0.01; Figure 6b). This indicated that ZHX2 interacts with the Notch1 promoter.
ZHX2 represses Notch1 promoter activity via NF-YA. (a) HepG2 cells were transfected with increasing amounts of ZHX2 and BEL7402 cells were transfected with increasing amounts of siZHX2, and the relative expression of Notch1 was quantified. (b) HepG2 and BEL7402 cells were co-transfected with pGL3- Notch1 along with pcDNA3.1 or increasing amounts of ZHX2, and the relative luciferase activity was measured, respectively. (c) Luciferase assay of the putative Notch1 promoter in HepG2 cells co-transfected with NF-YA or pcDNA3.1 or ZHX2. (d) ChIP analysis showed that ZHX2 repressed NF-YA occupying on the putative Notch1 promoter in HepG2 cells. All experiments were performed in triplicate. *P<0.05, **P<0.01
Interestingly, putative NF-YA binding sites were located upstream from Notch1 via Alggen Promo and JASPAR. ZHX2 functions as a repressor by interacting with NF-YA in HepG2 cells. To examine whether ZHX2 regulates Notch1 promoter activity indirectly, ZHX2, NF-YA, and wild-type promoter pGL3-Notch1 were co-transfected into HepG2 cells. We observed that ZHX2 could reduce the luciferase activity of NF-YA binding to wild-type Notch1 promoter, indicating that ZHX2 regulates Notch1 promoter activity by interacting with NF-YA (P<0.01; Figure 6c). ChIP assay showed that NF-YA was recruited to the Notch1 promoter fragment, and ZHX2 decreased the promoter occupancy of NF-YA (P<0.01; Figure 6d). Taken together, ZHX2 represses Notch1 transcription by interacting with NF-YA.
Discussion
Accumulating evidence revealed that miRNAs are pivotal mediators of HBV infections, hepatocarcinogenesis, and progression.14, 15 To our knowledge, the molecular details of HBV-related HCC patients with a family history of HCC are still largely elusive. In our previous study, through the comparison of miRNA expression profiles in the HBV-related HCC patients who had a family history of HCC with that in healthy volunteer, microarray results identified six miRNAs, miR-215, miR-3188, miR-374a, miR-215, miR-23b, miR-138-2*, and miR-216b, with over a 100-fold change in expression. Of the identified miRNAs, we focused on miR-3188 as its role has not been reported previously in HCC. Our data revealed that miR-3188 expression was markedly upregulated in HCC tissues compared with adjacent liver tissues and was significantly correlated with serum HBV DNA level, AFP expression, and prognosis after liver resection for HCC. Thus, the markedly overexpressed miR-3188 may play a pivotal role in the pathogenesis and progression of HCC.
CRISPR/Cas9 system is emerging as a state-of-art genome editing tool, and has a major impact on functional genetics in biological research.16 It is a microbial adaptive immune system composed of cas9 endonuclease, a sgRNA, and a transactivating CRISPR RNA (tracrRNA) that conferring targeted gene editing by small RNAs that guide the Cas9 nuclease to cleave foreign genetic elements.17 Recent studies reported that CRISPR/Cas9 could knock out miRNA expression more conveniently and efficiently.18 We designed CRISPR/Cas9 to target the sequences adjacent to Drosha processing sites in the secondary stem-loop structure of miR-3188, using the methodology presented by Chang et al.19 It has been reported that the inhibition of Drosha can lead to upregulation of primary miRNA (pri-miRNA) and in turn downregulation of mature miRNA.20 As expected, the accumulation of pri-miR-3188 and the downregulation of miR-3188 was found in the CRISPR/cas9-transfected cells. Then, we detected that miR-3188 KO suppresses cell growth, migration, and invasion of HCC cells in vitro and inhibits tumor growth and metastasis in vivo. All the evidences revealed that oncogene miR-3188 emerged as a potential therapeutic target and may behave as a tumor marker for the early detection of HBV-related hepatocarcinogenesis with a family history of HCC.
HBV infection is a major risk factor for HCC. Increasing evidence indicates an important role of HBV, especially HBx, in modulating the expression of cellular miRNAs that likely contribute to the pathogenesis of HCC. Xu et al.21 reported that HBx suppresses p53-mediated activation of miR-148a and activates mTOR signaling pathway through restoration of HPIP to enhance tumorigenesis. Wang et al.22 found that HBx-mediated downregulation of let-7a and upregulation of STAT3 leads to hepatocarcinogenesis. We also demonstrated that HBx suppresses p53-mediated activation of miR-216b, promotes IGF2BP2, and activates downstream signaling pathways in our previous study.13 In the present study, we found that overexpression of miR-3188 was positively correlated with serum HBV DNA level, and miR-3188 expression was also found upregulated in the liver tissues of HBV transgenic mice and HepG2.215 cells compared with C57BL/6 mice and HepG2 cells. Therefore, we further explored the underling mechanisms of HBV in regulating miR-3188 expression. We found that HBx was the key mediator of miR-3188 expression via transcriptional regulation. It has been reported that ectopic expression of HBx functions as a transactivating protein through interaction with transcription factors such as activating protein-1 (AP-1), NF-κB, and CREB.23 As expected, our data showed that HBx promotes miR-3188 expression via CREB-mediated activation.
Complicating the function of miRNAs in HBV-related HCC, it has been established that aberrantly expressed miRNAs can affect crucial signaling pathways. Our data demonstrated that miR-3188 KO can significantly inhibit the growth and metastasis of HCC cell lines in vitro and in vivo by inactivating Notch signaling pathways through upregulation of ZHX2. Mounting evidence confirmed that ZHX2 functions as a tumor suppressor and has been regarded as a transcriptional repressor negatively regulating the tumor markers AFP, GPC3, and H19.24 Yue et al.25 reported that ZHX2 could inhibit proliferation of HCC through transcriptionally regulating the expression of Cyclins A and E. Ma et al.26 found that ZHX2 represses transcription of genes associated with HCC by interfering with NF-YA in liver tumor cells. It has been demonstrated that HBx activates Notch signaling in HCC by promoting miR-3188 and repressing ZHX2 as described above. Thus, we presumed that ZHX2 might repress the Notch1 expression and inactivate Notch signaling in HCC by interacting with NF-YA. Interestingly, we found that ZHX2 indeed repressed Notch1 transcription by interacting with NF-YA. Taken together, the data proved that HBx promotes CREB-mediated activation of miR-3188 and activates Notch signaling through repressing ZHX2. Kongkavitoon et al.27 recently found that HBx activates Notch signaling via Delta-Like 4/Notch1 in HCC. However, the exact molecular mechanisms underlying Notch signaling functions and regulation need further investigation.
To our knowledge, the HBx–miR-3188–ZHX2–Notch1 signaling pathway regulation axis (Supplementary Figure 2f) was reported for the first time that may underlie the mechanisms of HBV-related HCC with a family history of HCC. MiR-3188 may be a useful diagnostic tool, a prognostic marker, and potential therapeutic target molecule that provides novel insights into HBV-related HCC. Further studies will be required to investigate whether detecting miR-3188 in plasma can be used to refine HCC surveillance guidelines based on the status of family history of HCC personalized for HBV patients.
Materials and Methods
Patients and specimens
A total of 150 pairs of human HCC tissues and their corresponding adjacent liver tissues (2 cm away from the tumor margin) were collected from patients who underwent liver resection between January 2007 and December 2008, and blood samples from 10 patients with HBV-related HCC and 10 healthy volunteers were collected between October and November 2015 at the Hepatic Surgery Center of Tongji Hospital affiliated with Huazhong University of Science and Technology (HUST). The follow-up was completed in December 2015. Written consent was obtained from all the patients and healthy volunteers before participating in the study. The study was approved by the medical ethics committee of Tongji Hospital, HUST, China.
CRISPR/Cas9 plasmid construction
The transfer plasmid lentiCRISPR v2 (#52961), packaging plasmids pCMV-VSV-G (#8454), and psPAX2 (#12260) was obtained from Addgene. The sgRNAs targeting hsa-miR-3188 were designed by CRISPR DESIGN (http://crispr.mit.edu/) that was developed by Dr. Feng Zhang’s Lab at MIT. The sgRNA-miR-3188 sequence is 5′-TCCTGCTCTGCTGTGCCGCC-3′. Specific target sequences were amplified and subcloned into the lentiCRISPR v2 plasmid and verified by DNA sequence analysis. The lentiCRISPR v2 plasmid encoding a nonspecific 20 nt guide RNA was used as a negative control for CRISPR/Cas9 knockout plasmids.
T7EN1 analysis
HepG2 cells were transfected with lentiCRISPR v2 containing sgRNA that targets miR-3188 or control plasmid. The genomic DNA was isolated for PCR amplification. The amplified fragments were purified using Genomic DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA). After being denatured and reannealed, the PCR products were digested with T7 endonuclease I (NEB). Digested fragments were separated on a 2% agarose gel. Images were captured by ChemiDoc XRS (Bio-Rad, Hercules, CA, USA).
Luciferase reporter assay
To verify the target sites of miRNAs, HepG2 and SMMC7721 cells were co-transfected with miR-3188 and luciferase reporter constructs containing wild-type or mutated ZHX2 3′-UTR. To study the effect of CREB on miR-3188 promoter activity, the plasmids containing fragments of human miR-3188 upstream region were respectively co-transfected with transfected with CREB or empty vector into HepG2 and SMMC7721 cells. To study the effect of NF-YA on Notch1 promoter activity, plasmids containing fragments of wild type or mutant NF-YA binding site were respectively co-transfected HepG2 cells with NF-YA, siZHX2, or empty vector into HepG2 cells. Cells were split and the activities were measured 24 h after transfection using Dual-Luciferase Assay Kit (Promega, Madison, WI, USA).
ChIP assays
ChIP assay was performed using Magna ChIP Assay Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. DNAs were coimmunoprecipitated with normal IgG, anti-CREB, or anti-NF-YA antibodies (Abcam, Cambridge, MA, USA) at 4 °C in a shaker overnight. MiR-3188 and Notch1 promoter was amplified by PCR using the primer pairs listed in Supplementary Table 1.
Statistical analysis
All the results were expressed as mean±S.D Categorical variables were compared using the χ2 test, Fisher’s exact test, or one-way ANOVA with P<0.05 considered statistically significant. OS was calculated by the Kaplan–Meier method and compared between the groups using the log-rank test. Multivariate analysis based on the proportional hazards model was used to identify the independent prognostic factors influencing OS. All the experimental data were analyzed using SPSS statistical software (version 16.0; SPSS Inc., Chicago, IL, USA).
References
El-Serag HB . Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012; 142: e1261.
Beasley RP, Hwang LY, Lin CC, Chien CS . Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet 1981; 2: 1129–1133.
Huang YT, Jen CL, Yang HI, Lee MH, Su J, Lu SN et al. Lifetime risk and sex difference of hepatocellular carcinoma among patients with chronic hepatitis B and C. J Clin Oncol 2011; 29: 3643–3650.
Bruix J, Sherman M . Management of hepatocellular carcinoma. Hepatology 2005; 42: 1208–1236.
Loomba R, Liu J, Yang HI, Lee MH, Lu SN, Wang LY et al. Synergistic effects of family history of hepatocellular carcinoma and hepatitis B virus infection on risk for incident hepatocellular carcinoma. Clin Gastroenterol Hepatol 2013; 11: e1631–e1633.
Yu MW, Chang HC, Chen PJ, Liu CJ, Liaw YF, Lin SM et al. Increased risk for hepatitis B-related liver cirrhosis in relatives of patients with hepatocellular carcinoma in northern Taiwan. Int J Epidemiol 2002; 31: 1008–1015.
Hassan MM, Spitz MR, Thomas MB, Curley SA, Patt YZ, Vauthey JN et al. The association of family history of liver cancer with hepatocellular carcinoma: a case-control study in the United States. J Hepatol 2009; 50: 334–341.
Volk ML, Lok AS . Is family history of liver cancer a risk factor for hepatocellular carcinoma? J Hepatol 2009; 50: 247–248.
Callegari E, Gramantieri L, Domenicali M, D'Abundo L, Sabbioni S, Negrini M . MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ 2015; 22: 46–57.
Xie KL, Zhang YG, Liu J, Zeng Y, Wu H . MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics 2014; 4: 1176–1192.
Gao P, Wong CC, Tung EK, Lee JM, Wong CM, Ng IO . Deregulation of microRNA expression occurs early and accumulates in early stages of HBV-associated multistep hepatocarcinogenesis. J Hepatol 2011; 54: 1177–1184.
Li Z, Rana TM . Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 2014; 13: 622–638.
Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang EL, Wu ZB et al. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis 2015; 6: e1670.
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 2011; 29: 4781–4788.
Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 2009; 49: 1098–1112.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823.
Hsu PD, Lander ES, Zhang F . Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014; 157: 1262–1278.
Jing W, Zhang X, Sun W, Hou X, Yao Z, Zhu Y . CRISPR/CAS9-mediated genome editing of miRNA-155 inhibits proinflammatory cytokine production by RAW264.7 Cells. Biomed Res Int 2015; 2015: 326042.
Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y . CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep 2016; 6: 22312.
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003; 425: 415–419.
Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest 2013; 123: 630–645.
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P et al. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol 2010; 53: 57–66.
Diao J, Garces R, Richardson CD . X protein of hepatitis B virus modulates cytokine and growth factor related signal transduction pathways during the course of viral infections and hepatocarcinogenesis. Cytokine Growth Factor Rev 2001; 12: 189–205.
Morford LA, Davis C, Jin L, Dobierzewska A, Peterson ML, Spear BT . The oncofetal gene glypican 3 is regulated in the postnatal liver by zinc fingers and homeoboxes 2 and in the regenerating liver by alpha-fetoprotein regulator 2. Hepatology 2007; 46: 1541–1547.
Yue X, Zhang Z, Liang X, Gao L, Zhang X, Zhao D et al. Zinc fingers and homeoboxes 2 inhibits hepatocellular carcinoma cell proliferation and represses expression of Cyclins A and E. Gastroenterology 2012; 142: e1552.
Ma H, Yue X, Gao L, Liang X, Yan W, Zhang Z et al. ZHX2 enhances the cytotoxicity of chemotherapeutic drugs in liver tumor cells by repressing MDR1 via interfering with NF-YA. Oncotarget 2015; 6: 1049–1063.
Kongkavitoon P, Tangkijvanich P, Hirankarn N, Palaga T . Hepatitis B virus HBx activates Notch Signaling via Delta-Like 4/Notch1 in hepatocellular carcinoma. PLoS ONE 2016; 11: e0146696.
Acknowledgements
We thank all the authors for discussing the project. This work was supported by the National Natural Science Foundation of China (81202300 to Hui-Fang Liang).
Author contributions
Conception and design of the study: Fu-Yao Liu, Yue-Ling Deng, and Hui-Fang Liang; generation, collection, assembly, analysis, and/or interpretation of data: Fu-Yao Liu and Shao-Jun Zhou; drafting or revision of the manuscript: Fu-Yao Liu, Shao-Jun Zhou, and Jonathan C Jaoude; approval of the final version of the manuscript: all authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by H-U Simon
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Zhou, Sj., Deng, Yl., Liang, Hf. et al. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ 24, 1577–1587 (2017). https://doi.org/10.1038/cdd.2017.87
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2017.87
- Springer Nature Limited