Abstract
Background:
The cyclin-dependent kinase inhibitor 3 (CDKN3) has been perceived as a tumour suppressor. Paradoxically, CDKN3 is often overexpressed in human cancer. It was unclear if CDKN3 overexpression is linked to alternative splicing variants or mutations that produce dominant-negative CDKN3.
Methods:
We analysed CDKN3 expression and its association with patient survival in three cohorts of lung adenocarcinoma. We also examined CDKN3 mutations in the Cancer Genome Atlas (TCGA) and the Moffitt Cancer Center’s Total Cancer Care (TCC) projects. CDKN3 transcripts were further analysed in a panel of cell lines and lung adenocarcinoma tissues. CDKN3 mRNA and protein levels in different cell cycle phases were examined.
Results:
CDKN3 is overexpressed in non small cell lung cancer. High CDKN3 expression is associated with poor overall survival in lung adenocarcinoma. Two CDKN3 transcripts were detected in all samples. These CDKN3 transcripts represent the full length CDKN3 mRNA and a normal transcript lacking exon 2, which encodes an out of frame 23-amino acid peptide with little homology to CDKN3. CDKN3 mutations were found to be very rare. CDKN3 mRNA and protein were elevated during the mitosis phase of cell cycle.
Conclusions:
CDKN3 overexpression is prognostic of poor overall survival in lung adenocarcinoma. CDKN3 overexpression in lung adenocarcinoma is not attributed to alternative splicing or mutation but is likely due to increased mitotic activity, arguing against CDKN3 as a tumour suppressor.
Similar content being viewed by others
Main
The human CDKN3 gene encodes the cyclin-dependent kinase inhibitor 3, which is a dual specificity protein tyrosine phosphatase of the CDC14 group. CDKN3 binds cyclin-dependent kinases CDK1 and CDK2 (Hannon et al, 1994) and dephosphorylates their activating loop Thr residues (Poon and Hunter, 1995; Brown et al, 1999; Song et al, 2001). Dephosphoryation of the activating Thr site reduces substrate and ATP binding activities of the CDKs (Brown et al, 1999). Thus, CDKN3 is a negative regulator of CDK1 and CDK2.
Since CDK-driven cell cycle is essential for proliferation of cancer cells and CDKN3 inhibits CDK activities, CDKN3 has been perceived as a tumour suppressor (Yeh et al, 2000; Yu et al, 2007; Nalepa et al, 2013). Paradoxically, CDKN3 is often overexpressed in various human cancer tissues and cancer cell lines (Lee et al, 2000; Yu et al, 2007; Espinosa et al, 2013). Lee et al (2000) found overexpression of CDKN3 mRNA and protein in breast and prostate cancer. Inhibition of CDKN3 expression by antisense CDKN3 suppressed anchorage-independent colony formation in vitro and tumour xenograft growth in vivo (Lee et al, 2000), suggesting that increased CDKN3 expression in cancer cells plays a positive role in the transformed phenotypes. Yu et al (2007) found CDKN3 mRNA overexpression in astrocytoma and that elevated CDKN3 mRNA level was significantly associated with shorter patient survival. More recently, Espinosa et al (2013) identified CDKN3 as one of the six most upregulated mitosis pathway genes in cervical cancer. Moreover, CDKN3 overexpression is linked to the poor survival of these cervical cancer patients (Espinosa et al, 2013).
However, based on the perception that CDKN3 is a potential tumour suppressor, a possible explanation of CDKN3 mRNA overexpression in cancer is the presence of dominant-negative CDKN3 mutations, which could not be distinguished from the normal CDKN3-encoding mRNA in the gene expression data. Hence, CDKN3 overexpression may actually result in overexpression of dominant-negative CDKN3 mutants. It was reported that aberrant CDKN3 transcripts from alternated splicing, insertion/deletion and nonsense mutations were found in hepatocellular carcinoma (Yeh et al, 2000; Yeh et al, 2003). In another study (Yu et al, 2007), two CDKN3 transcripts (variant a and c) were identified in all brain samples and two additional transcripts (variants b and d) were found in some glioblastoma samples. Variant a encodes the full length 212-amino acid CDKN3. Variants b–c resulted from alternative splicing around exons 2–3. Variant c is an exon 2 skip transcript that results in a short 23-amino acid peptide with little sequence homology to CDKN3. Variant b results in a short 8-amino acid peptide. Variant d lacks a part of exon 2 and the entire exon 3, resulting in a 179-amino acid protein. None of variant b–c products has been documented to have any biological activity. Thus, whether CDKN3 gene overexpression in cancer is linked to alternative splicing or mutations that generate dominant-negative CDKN3 remains to be further assessed.
Alternatively, the concept that CDKN3 is a tumour suppressor may be inaccurate. While CDKs drive cell cycle, CDK activities oscillate along the cell cycle. This involves not only CDK activation but also CDK deactivation. Indeed, constitutive CDK2 activation by preventing Thr14/Tyr15 phosphorylation causes problems in DNA replication and genome integrity (Hughes et al, 2013). A decrease in CDK1-cyclin B activity is required for mitotic exit (Wurzenberger and Gerlich, 2011; Nalepa et al, 2013). Thus, decreasing CDK activity may be just as important as increasing CDK activity in coordinated regulation of cell cycle. Consistent with this notion, knockdown of CDKN3 leads to mitotic failure (Nalepa et al, 2013). Therefore, an alternative explanation of CDKN3 overexpression in human cancer is that CDKN3 is important for the safe passage through cell cycle and the elevated CDKN3 level is attributed to the higher mitotic activity in cancer cells.
In this study, we present evidence that CDKN3 is overexpressed in human NSCLC and higher CDKN3 expression level is associated with poor survival of lung adenocarcinoma patients. We detected the full length and the exon 2 skip CDKN3 transcripts in all cell lines and lung tumour tissues and found no evidence of change in alternative splicing. Furthermore, CDKN3 mutations are rare in TCGA tumours and in the TCC data set of 3383 tumours. Interestingly, we found that CDKN3 expression is upregulated during the mitosis (M)-phase of cell cycle in cultured cells.
Materials and Methods
Reagents, cells and tissue samples
Monoclonal anti-CDKN3 antibody 2H10 was purchased from Abcam (Cambridge, MA, USA). Monoclonal antibody No. 61033410 was from BD Biosciences (San Jose, CA, USA). Rabbit monoclonal antibody (D2C8) to phospho-histone H3 Ser10 (pHH3) was from Cell Signaling Technology (Danvers, MA, USA). Anti-actin antibody was obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Thymidine and nocodazole were from Sigma (St Louis, MO, USA).
Lung cancer cell line and hTBE/v cells have been maintained in a central repository at the Moffitt Cancer Center since 2008. All cell lines in the Moffitt repository had been authenticated by STR analysis (ACTG Inc, Wheling, IL, USA) as of September 2010, and all cells had been routinely tested and were negative for mycoplasma (PlasmoTest, InvivoGen, San Diego, CA, USA). BEAS2B cells were from American Type Culture Collection and cultured according to the supplier’s instruction. The primary human endothelium (HUVEC) cells were obtained from Sarah Yuan at the University of South Florida. Other cell lines were from the laboratories of Jin Cheng or Kenji Fukasawa at the Moffitt Cancer Center or have been reported (Ren et al, 2010; Ren et al, 2013).
Patient data and samples
All work was approved by the University of South Florida Institutional Review Board. Gene expression and survival data from a cohort of 398 lung cancer patients diagnosed with adenocarcinoma recruited from Moffitt Cancer Center’s TCC (Fenstermacher et al, 2011) between April 2006 and August 2010 were included in this study. This large lung adenocarcinoma cohort has been extensively characterised by Moffitt’s Lung Cancer Center for Excellence will be described in detail elsewhere. Briefly, RNA isolated from fresh frozen lung tumours were profiled using a custom Affymetrix GeneChip (Affymetrix, Santa Clara, CA, USA) that measured the expression of ∼60 000 distinct transcripts. CEL files were normalised against their median sample using IRON (Welsh et al, 2013). An RNA-quality-related batch effect was identified in the resulting normalised data, which was removed by training a partial least squares (PLS) model (Wold et al, 1984) to the BioAnalyzer BA_RIN RNA quality metric (Agilent Technologies, Santa Clara, CA, USA), then subtracting the first PLS component. Survival analyses were performed to determine if CDKN3 expression levels were associated with overall survival using Kaplan–Meier survival curves and the log-rank test. Overall survival was right-censored at 5 years and was calculated from the date of diagnosis until the date of last follow-up or death.
RNA was isolated from six de-identified RNA samples from this cohort RNA from freshly frozen tumour tissue macrodissected to contain >70% tumour tissue and RNA was extracted using the manufacturer’s instructions for the Qiagen RNAeasy Kit (Valencia, CA, USA) by Moffitt’s Tissue Core. The isolated RNA was quantified using Nanodrop (Thermo Scientific, Wilmington, DE, USA) and Qubit instrument (ThermoFisher Scientific, Waltham, MA, USA).
Plasmids
A CDKN3 lentiviral expression vector was constructed by cloning the CDKN3 coding sequence (identical to CDKN3 coding sequence of Genebank entry NM005192) from HCC827 cells into Pme I site of the pWPI lentiviral vector obtained from Addgene (Cambridge, MA, USA). pLKO.1-based CDKN3 shRNA lentiviral vectors TRCN0000002525 and TRCN0000002527, and a non-target shRNA were obtained from Sigma. Lentiviruses were prepared similar to that described previously (Ren et al, 2010).
RT–PCR and RT–qPCR
Total RNA from cells were extracted using Trizol reagent (Life Technologies, Benicia, CA, USA). RT–PCR was performed using SuperScript One-Step RT–PCR Platinum Taq system (Life Technologies) with 50 ng RNA and one of the following primer pairs CK1/CK1R, CK2/CK2R, CK3/CK3R or CK1/CK3R (See Supplementary Information). RT–PCR products were separated on agarose gel, purified and re-amplified by PCR. PCR products were cleaned with ExoSAP-IT PCR product cleanup reagent (Affymetrix) and sequenced in both strains using BigDye Terminator 3.1 cycle sequencing kit (Life Technologies) as described (Ren et al, 2013). Quantitative RT–PCR (RT–qPCR) was performed using CK2/CK2R or CK-T/CK-R primer pairs (Supplementary Information). RT–qPCR was performed using Power SYBR Green reagents (Applied Biosystems, Foster City, CA, USA). Samples were assayed in triplicates as described (Schneeberger et al, 2014).
Cell culture, synchronization and immunoblotting
Lung cell lines A549, H2172, H3122, H2228, H1975, HCC827, H3255 and BEAS2B were cultured in RPMI 1640/10% foetal bovine serum (FBS). HeLa, PC-3, PANC-1, U2OS, HCA2, SW480, MCF7 and HepG2 cell lines were cultured in Dulbecco’s modified Eagle’s medium/10% FBS. HUVEC cells were cultured in endothelial cell growth medium (Lonza, Allendale, NJ, USA). hTBE/v cells were cultured in BEBM with additives (Lonza). Cells were maintained in 5% CO2 humidified atmosphere at 37 °C.
To enrich cells in G0/early G1 phase, cells were serum starved for 48 h. For double thymidine block to enrich cells in later G1/S phase, cells (30% confluent) were treated with 2 mM thymidine for 18 h. The cells were washed with PBS, incubated with normal cell culture medium for 9 h and then treated with 2 mM thymidine for another 16 h. For nocodazole blockage to enrich M phase cells, cells (80–90% confluent) were treated with 100 ng ml−1 nocodazole for 12 h. After removing medium, round and loosely attached mitotic cells were dislodged by shaking, suspended in medium and collected by low speed centrifugation.
Cell lysates were prepared similar to that described (Ren et al, 2013; Schneeberger et al, 2014; Schneeberger et al, 2015) by lysis in cold lysis buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 25 mM NaF, 5 mM sodium pyrophosphate, 1 mM Na3VO4, 2 μg ml−1 aprotinin, 2 μg ml−1 leupeptin, 100 μg ml−1 phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 20 mM p-nitrophenyl phosphate, 1% Triton X-100). Cell lysate supernatants were obtained by microcentrifugation (16 000 r.p.m., 10 min at 4 °C). Equal amounts of proteins were separated on 12 or 15% SDS-polyacrylamide gels, transferred to nitrocellulose filters and analysed by immunoblotting with indicated antibodies.
CDKN3 knockdown and cell proliferation assay
Lentiviruses were prepared as described (Ren et al, 2010) and used to infect A549 and HeLa cells. Infected cells were selected by puromycin. Puromycin-resistant cells were pooled and analysed. Cell proliferation was assayed as described (Ren et al, 2010) in 96-well plate in triplicates. Relative viable cells were measured using Cell TiterGlo reagent (Promega, Madison, WI, USA). Statistical analysis was performed using the nonparametric Mann–Whitney test. P<0.05 was considered significantly different.
Results
CDKN3 is overexpressed in NSCLC
By examination of public databases, we noticed that CDKN3 is often overexpressed in various tumours (Supplementary Figure 1), including lung cancer. To explore the expression of CDKN3 mRNA in NSCLC, we first compared CDKN3 expression in NSCLC tumours and matched normal tissues using data from GSE19188 (Hou et al, 2010) as previously described (Engel et al, 2013). The Affymetrix U133 plus 2.0 microarrays (Affymetrix) data in GSE19188 contain 87 tumour samples and 58 adjacent normal tissue samples (Figure 1A). The log2 CDKN3 expression level was 4.345±0.534 (normal tissues, mean±s.d.), 7.048±1.104 (adenocarcinoma), 7.617±1.461 (large cell carcinoma) and 8.000±0.683 (squamous cell carcinoma). Thus, CDKN3 expression was significantly elevated in all three histological subtypes of NSCLC (P<0.0001). We then expanded the analysis to a larger cohort of TCGA RNA-seq expression data that contain 490 lung adenocarcinoma, 491 lung squamous cell carcinoma and 108 of their matched normal tissues (Figure 1B). Again, CDKN3 expression in both adenocarcinoma and squamous cell carcinoma was statistically (P<0.0001) higher than in the matched normal tissues.
CDKN3 expression is elevated in NSCLC. (A) CDKN3 expression in the GSE19188 data set of NSCLC tumour and available adjacent normal samples (Engel et al, 2013). LUAD, lung adenocarcinoma. LULC, lung large cell carcinoma. LUSC, lung squamous cell carcinoma. Gene expression was determined via microarray for CDKN3 using probe 209714_s_at. The horizontal lines are the mean+s.d. of each group of samples. (B) CDKN3 expression in LUAD, LUSC and matched normal tissues from TCGA. The normal group is the combined data from the matched normal of LUAD and LUSC. The difference between the matched normal of LUAD and LUSC was not statistically significance.
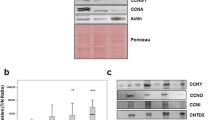
It was reported that CDKN3 mRNA and protein were overexpressed in breast and prostate cancer cell lines (Lee et al, 2000). To determine if CDKN3 expression is also elevated in lung cancer cell lines, we performed RT–qPCR analysis of CDKN3 mRNA in the immortalised human lung epithelial cell line (hTBE/v) and six lung adenocarcinoma cell lines. Compared with hTBE/v cells, higher levels of CDKN3 mRNA were detected in these lung cancer cell lines (Figure 2A).
Expression of CDKN3 mRNA and protein in lung cell lines. (A) RNA was isolated from indicated cells and the relative amounts of CDKN3 mRNA were determined by RT–qPCR using primers CK2/CK2R. The CDKN3 mRNA level in hTBE/v cells was arbitrary set as 1. (B) HEK293 cells were transfected with empty or CDKN3-encoding lentiviral vector (plasmid). Cell lysates from non-transfected and transfected cells were analysed with antibodies 2H10 or actin. NS, a non-specific reactive band. (C) Cell lysates from indicated cells were analysed by immunoblotting with CDKN3 antibody 2H10.
The identity of CDKN3 protein on immunoblots has been questioned (Yu et al, 2007). To correctly identify CDKN3 protein, we cloned the CDKN3 protein coding sequence from HCC827 cells and expressed the non-tagged, 212-amino acid CDKN3 protein coding cDNA (identical to NM_005192.3) in HEK293 cells. Cell lysates from non-transfected cells, empty vector-transfected cells and CDKN3 expression vector-transfected cells were analysed with an anti-CDKN3 mouse monoclonal antibody 2H10 (Figure 2B) or another mouse monoclonal antibody (BD cat. No. 610334, Supplementary Figure 2). Antibody 2H10 correctly detected CDKN3 at 27 kDa and also reacted with a non-specific bands at 23 kDa. Antibody No. 610334 reacted with a 34-kDa band in all three cell lysates (Supplementary Figure 2). Thus, the 27-kDa band reacting with antibody 2H10 is CDKN3. Additional evidence to support the identification of the 27-kDa band as CDKN3 was obtained by knocking down endogenous CDKN3 in cell lines with shRNAs (see below). After establishing the identity of CDKN3 in immunoblots, we probed lung cell lysates for CDKN3 protein. Figure 2C shows that higher levels of CDKN3 protein were present in lung cancer cell lines.
CDKN3 overexpression is associated with poor overall survival in lung adenocarcinoma
We first analysed two public lung adenocarcinoma databases to determine the relationship between CDKN3 expression in tumours and patients outcome in terms of overall survival. The microarray-based Molecular Classification of Lung Adenocarcinoma (MCLA) from the Director’s Challenge Consortium contains 478 patients (Shedden et al, 2008) (Figure 3A). The RNA-seq-based lung adenocarcinoma cohort from TCGA contains 381 patients (Figure 3B). In both cohorts, CDKN3 expression was prognostic. The overall survival rates for patients with high CDKN3 levels (above the median) were significantly lower than those with low CDKN3 levels (below the median (P-values of 0.0046 and 0.0059, respectively)). We also examined the 390 patient lung squamous cell carcinoma cohort from TCGA. Unlike the lung adenocarcinoma cohorts, CDKN3 expression was not significantly prognostic in the TCGA lung squamous cell carcinoma cohort (Figure 3C). However, comparing with lung adenocarcinoma, lung squamous cell carcinoma has an overall significantly (P<0.0001) higher CDKN3 transcript level (Figure 1). Thus, the ‘low CDKN3’ expression levels in lung squamous cell carcinoma are very high already.
CDKN3 expression is highly prognostic in three lung adenocarcinoma cohorts. CDKN3 mRNA level was correlated with patient outcome by dichotomizing patients into low and high expression groups based on the median expression value. The overall survival of the two CDKN3 groups was compared by Kaplan–Meier estimate and two-sided log-rank test. CDKN3 mRNA and patient survival data were from (A) MCLA, (B, C) TCGA, (D) Moffitt's Lung SPORE 422.
Next, we examined CDKN3 expression in a cohort of 398 lung adenocarcinomas developed at the Moffitt Cancer Center. Figure 3D shows that the Lung SPORE422 patients with high CDKN3 levels had significantly shorter overall survival than those with lower CDKN3 levels (P<0.0006). No significant differences in CDKN3 mRNA levels were found among different stages of lung adenocarcinoma (Supplementary Figure 3A and B). Because more of stage I patient survival and CDKN3 mRNA data were available than other stages of lung adenocarcinoma, we next analysed those stage I patients whose survival data are available (n=254). Again, among stage I patients, those with high CDKN3 levels had a shorter survival probability (Supplementary Fig. 3C). Taken together, these data indicate that high CDKN3 levels are associated with poor overall survival in lung adenocarcinoma patients.
Two CDKN3 transcripts are present in normal and tumour cells and lung tumour tissues
We designed RT–PCR primer pairs CK1/CK1R located in exons 1 and 5 (Figure 4) that allowed us to detect both the full length CDKN3 transcript and splicing variants between exons 2–5 reported previously by other investigators (Yeh et al, 2000; Yu et al, 2007). RNA from primary HUVEC cells, 3 non-transformed, immortalised cell lines and 14 cancer cell lines were prepared (Table 1). The CDKN3 transcripts were amplified by RT–PCR. Two RT–PCR products were detected in all samples (Figure 4 and Supplementary Figure 4). These two PCR products were isolated from gels, re-amplified by PCR using the same primers and then sequenced. cDNA sequencing data showed that both the long and the short PCR fragments from all samples were identical (Figure 4, Table 1, and Supplementary Figure 5). The long form is the CDKN3-encoding mRNA identical to NM_005192.3. The short RT–PCR product from the CK1/CK1R primer pairs yields a 284-bp fragment resulting from exon 2 skip (equivalent to variant c reported in the study by Yu et al (2007), Figure 4). This Exon 2 skip variant results in a frameshift that is predicted to encode a 23-amino acid peptide with little sequence homology to CDKN3 (Figure 4 and Supplementary Figure 5). No alternative splicing variant in the boundary of exon 2 was detected by cDNA sequencing. No other smaller RT–PCR product, as would be predicted from a longer excision of the mRNA by alternative splicing, was detected. Also, no point mutation was detected. To further evaluate the possibility of the existence of other alternative splicing variants, we used primer pairs CK1/CK3 that cover the entire coding sequence to do RT–PCR. Again, we detected only two PCR products that correspond to the CDKN3 and the exon 2 skip form. To explore the possibility of point mutation in regions not covered by CK1/CK3 primers, we performed RT–PCR using primer pairs CK2/CK2R and CK3/CK3R (Figure 4), both of them yield a single, predicted PCR product in all samples. No mutation was found in the sequencing data of these PCR products. Next, we obtained mRNAs from six lung adenocarcinoma tissues. The mRNAs from these tissue samples were analysed as above. Data from these lung tumour samples were identical to those obtained from cell lines (Table 1). Taken together, these results show that two CDKN3 transcripts are present in cells and tissues that we have analysed. No mutation or alternative splicing is found in our cohort of cancer cells and tissues.
Schematic presentation of CDKN3 transcripts, exon boundaries and PCR primers. The CDKN3 mRNA (Genebank NM_005192.3) is shown. Heavy box indicates the protein coding sequence. Exons 1–8 are according to the notation of Genebank NM_005192.3. The amino acid sequence encoded by the short (exon 2 skip) splicing variant is shown and aligned with the long, CDKN3-encoding transcript. fs, frame shift. Also shown are locations of three pairs of PCR primers.
CDKN3 gene mutations are rare in human cancer
To further assess the possibility of CDKN3 mutations in human cancer, we examined the massively parallel sequencing data set of 3383 tumour tissues from 48 tumour types in the TCC project (Fenstermacher et al, 2011; Ren et al, 2013). Twenty-three CDKN3 mutation cases were found in 11 tumour types (Table 2). No disruptive changes (frameshift insertion/deletion, nonsense mutation, splice site mutation) were found. Of these 23 mutations, 13 mutations at 3 unique positions of Q8R, I108V and T127I were observed in either 1000 Genomes (Genomes Project C et al, 2012) or Exome Sequencing Project (Exome Variant Server, http://evs.gs.washington.edu/EVS/, accessed February 2015) of healthy people, and are likely inherited variants. They are all rare, and none were observed in the homozygous state. Therefore, few mutations are observed in this gene: at most 10 in 3383 individual across many tumour types. In addition, there are low levels (1–2%) of CDKN3 gene amplification in TCGA lung adenocarcinoma.
We next extend the examination to lung adenocarcinoma in the TCGA research network (Cancer Genome Atlas Research N, 2014) and the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al, 2012) data sets (www.cbioportal.org (Cerami et al, 2012; Gao et al, 2013)). No CDKN3 mutation was detected in lung adenocarcinoma tumour tissues in the TCGA cohort or in lung cancer cell lines in the CCLE cohort. Also, no CDKN3 mutation in lung cancer is found in the Broad Institute’s tumorportal database (www.tumorportal.org) (Lawrence et al, 2014). Thus, the frequent overexpression of CDKN3 in lung cancer is not associated with gene mutation.
Cells in the mitotic phase have high levels of CDKN3
To determine if CDKN3 expression fluctuates during the cell cycle, we synchronized HeLa and A549 cells in later G1/S phase by double thymidine blockage. Cells at various time points following release from the second thymidine block were collected. Cell lysates were analysed by immunoblotting with antibodies to CDKN3 and the mitotic-specific histone H3 Ser10 phosphorylation (pHH3) (Hendzel et al, 1997). As shown in Figure 5A and B, low levels of CDKN3 protein were present in double thymidine-blocked HeLa and A549 cells. The increase in CDKN3 protein precedes the appearance of pHH3 and peaked at the same time point as pHH3 did. We next extended the analysis to additional lung cancer cell lines as well as HeLa cells by blocking cells in G0/early G1 phase with serum deprivation, in later G1/S phase with double thymidine block and in M phase with nocodazole. Cell lysates were prepared from these cells and analysed by immunoblotting. As shown in Figure 5A–C, cells in the M phase consistently showed the highest CDKN3 protein level.
CDKN3 expression is elevated in M phase. (A and B, left panels) HeLa and A549 cells were subjected to double thymidine blockage. At the indicated time after release from the second thymidine block, cell lysates were collected and analysed by immunoblotting with indicated antibodies. (A and B, right panels, and C) Cell lysates from unsynchronized (U), serum-deprived (G0/1), double thymidine-blocked (S) and nocodazole block (M) cells were analysed by immunoblotting with indicated antibodies. (D) Cells were treated as in C. RNA was isolated and analysed by RT–qPCR using CK-T/CK-R primers. The data were averages from two experiments. The relative amount of CDKN3 mRNA in the serum-deprived cells from each cell line was set as 1.
To assess if the difference in CDKN3 protein level was related to the mRNA level, RT–qPCR was performed. Consistent with the results from protein analysis, serum-starved cells have the lowest level of CDKN3 mRNA, whereas cells in the M phase have high levels of CDKN3 although high level of CDKN3 mRNA was also detected in some cells blocked in the S phase (Figure 5D). Overall, the relative amount of CDKN3 protein during the three phases of cell cycles correlates with the mRNA level.
If CDKN3 expression is increased in the mitotic phase, it is predicted that CDKN3 expression would correlate with the expression of mitotic-related genes. Consistently, gene expression correlation data of lung adenocarcinoma from TCGA shows that genes with the highest expression correlation with CDKN3 are mitotic-related genes (Supplementary Table 1).
Knockdown of CDKN3 reduces cell proliferation
If CDKN3 plays a positive role in regulating mitotic cell cycle, reducing CDKN3 would be predicted to attenuate cell proliferation. Supplementary Figure 6 shows that we were able to knockdown CDKN3 in A549 and HeLa cells. CDKN3 knockdown in A549 and HeLa cells resulted in significantly reduced cell proliferation of these cells (Supplementary Figure 6).
Discussion
Following the initial observation of CDKN3 overexpression in various types of human cancer, we performed more detailed characterisation of CDKN3 expression in NSCLC. We found CDKN3 expression is elevated in NSCLC. Three cohorts of lung adenocarcinoma consisted of 1328 patients and used either mRNA microarray or RNA-seq method to measure CDKN3 expression; high CDKN3 expression is consistently associated with significantly shorter overall survival of these patients. No such association is found in lung squamous cell carcinoma. A possible explanation is that lung squamous cell carcinoma has an overall high level of CDKN3 expression. The overall high level of CDKN3 expression within lung squamous cell carcinoma may render indifference between ‘low’ and ‘high’ levels in the prognosis of CDKN3 for the overall survival in this histological type of lung cancer.
When this paper is under peer review, Zang et al (2015) reported a mega-analysis of data sets in Lung Cancer Explorer (http://qbrc.swmed.edu/lce/) database using different analytic methods. They also found that CDKN3 is upregulated in lung adenocarcinoma and lung squamous cell carcinoma and that lung squamous cell carcinoma has an overall higher level of CDKN3. Furthermore, the high CDKN3 levels are associated with poor survival in lung adenocarcinoma but not in lung squamous cell carcinoma. These findings are in agreement with the data that we presented here.
Based on the perception of CDKN3 as a potential tumour suppressor, a possible mechanistic explanation of CDKN3 overexpression in the tumours is alternative splicing or mutations that produce dominant-negative products of CDKN3. However, in our cohort of 24 samples, we found that all cells and tumours express the same 2 transcripts. We detected no aberrant splice variant or point mutation in the CDKN3 transcripts. A difference in our analysis and the previous reports in hepatocellular carcinoma and glioma is that previous studies performed sequencing analyses after cloning the cDNA (Yeh et al, 2000; Yu et al, 2007), whereas we sequenced the RT–PCR products directly. Sequencing analysis of cDNA clones is more sensitive to detect minute changes, although the origins of these changes are unknown. While less sensitive, data of our direct sequencing analyses of RT–PCR products are better representative of what is present as the whole in the cells and thus the overall functionality of CDKN3 overexpression in the cells. Similar to the results of Lee et al (2000), our experiments showed that knockdown of CDKN3 inhibited cell proliferation, suggesting that CDKN3 has a positive role in cell proliferation.
CDKN3 mutation data from TCC, TCGA, CCLE and tumorportal indicate that CDKN3 mutations are rare in human cancer. Importantly, no disruptive mutation is found. Recurrent missense Q8R, I108V and T127I mutations were found in TCC tumours. These changes were also present in healthy people and thus may represent polymorphic variants. It remains to be determined if these variants have any functional consequence. Nevertheless, the only two cases of T127I change found in lung tumours cannot account for the poor survival of high CDKN3 expression patients in approximately one-half of 1328 lung adenocarcinoma cases. Taken together, our analyses indicate that the functional CDKN3, not dominant-negative CDKN3 mutants, is overexpressed in lung adenocarcinoma.
Significantly, we found high level of CDKN3 expression in mitotic cells, whereas cell enriched in the G0/G1 phase by serum-starvation had the lowest level of CDKN3. Consistently, TCGA data show that CDKN3 has the highest expression correlation with genes involving in mitosis. The study by Espinosa et al (2013) also identified CDKN3 as a mitosis pathway gene in cervical cancer. Our finding of elevated CDKN3 in mitotic cells is in agreement with the notion that CDKN3 has an important role in mitosis (Nalepa et al, 2013) and suggest that CDKN3 should be added to the list of phosphatases important for cell cycle through mitosis (Wurzenberger and Gerlich, 2011). It also suggests that the high level of CDKN3 expression in human cancer is likely to reflect the increased fraction of mitotic cells in the tumours.
Change history
22 December 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483 (7391): 603–607.
Brown NR, Noble ME, Lawrie AM, Morris MC, Tunnah P, Divita G, Johnson LN, Endicott JA (1999) Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J Biol Chem 274 (13): 8746–8756.
Cancer Genome Atlas Research N (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511 (7511): 543–550.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2 (5): 401–404.
Engel BE, Welsh E, Emmons MF, Santiago-Cardona PG, Cress WD (2013) Expression of integrin alpha 10 is transcriptionally activated by pRb in mouse osteoblasts and is downregulated in multiple solid tumours. Cell Death Dis 4: e938.
Espinosa AM, Alfaro A, Roman-Basaure E, Guardado-Estrada M, Palma I, Serralde C, Medina I, Juarez E, Bermudez M, Marquez E, Borges-Ibanez M, Munoz-Cortez S, Alcantara-Vazquez A, Alonso P, Curiel-Valdez J, Kofman S, Villegas N, Berumen J (2013) Mitosis is a source of potential markers for screening and survival and therapeutic targets in cervical cancer. PloS One 8 (2): e55975.
Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS (2011) Implementing personalized medicine in a cancer center. Cancer J 17 (6): 528–536.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6 (269): pl1.
Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (2012) An integrated map of genetic variation from 1092 human genomes. Nature 491 (7422): 56–65.
Hannon GJ, Casso D, Beach D (1994) KAP: a dual specificity phosphatase that interacts with cyclin-dependent kinases. Proc Natl Acad Sci USA 91 (5): 1731–1735.
Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106 (6): 348–360.
Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S (2010) Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PloS One 5 (4): e10312.
Hughes BT, Sidorova J, Swanger J, Monnat RJ Jr, Clurman BE (2013) Essential role for Cdk2 inhibitory phosphorylation during replication stress revealed by a human Cdk2 knockin mutation. Proc Natl Acad Sci USA 110 (22): 8954–8959.
Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505 (7484): 495–501.
Lee SW, Reimer CL, Fang L, Iruela-Arispe ML, Aaronson SA (2000) Overexpression of kinase-associated phosphatase (KAP) in breast and prostate cancer and inhibition of the transformed phenotype by antisense KAP expression. Mol Cell Biol 20 (5): 1723–1732.
Nalepa G, Barnholtz-Sloan J, Enzor R, Dey D, He Y, Gehlhausen JR, Lehmann AS, Park SJ, Yang Y, Yang X, Chen S, Guan X, Chen Y, Renbarger J, Yang FC, Parada LF, Clapp W (2013) The tumour suppressor CDKN3 controls mitosis. J Cell Biol 201 (7): 997–1012.
Poon RY, Hunter T (1995) Dephosphorylation of Cdk2 Thr160 by the cyclin-dependent kinase-interacting phosphatase KAP in the absence of cyclin. Science 270 (5233): 90–93.
Ren Y, Chen Z, Chen L, Fang B, Win-Piazza H, Haura E, Koomen JM, Wu J (2010) Critical role of Shp2 in tumour growth involving regulation of c-Myc. Genes Cancer 1 (10): 994–1007.
Ren Y, Zhang Y, Liu RZ, Fenstermacher DA, Wright KL, Teer JK, Wu J (2013) JAK1 truncating mutations in gynaecologic cancer define new role of cancer-associated protein tyrosine kinase aberrations. Sci Rep 3: 3042.
Schneeberger VE, Luetteke N, Ren Y, Berns H, Chen L, Foroutan P, Martinez GV, Haura EB, Chen J, Coppola D, Wu J (2014) SHP2E76K mutant promotes lung tumorigenesis in transgenic mice. Carcinogenesis 35 (8): 1717–1725.
Schneeberger VE, Ren Y, Luetteke N, Huang Q, Chen L, Lawrence HR, Lawrence NJ, Haura EB, Koomen JM, Coppola D, Wu J (2015) Inhibition of Shp2 suppresses mutant EGFR-induced lung tumours in transgenic mouse model of lung adenocarcinoma. Oncotarget 6: 6191–6202.
Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, Chang AC, Zhu CQ, Strumpf D, Hanash S, Shepherd FA, Ding K, Seymour L, Naoki K, Pennell N, Weir B, Verhaak R, Ladd-Acosta C, Golub T, Gruidl M, Sharma A, Szoke J, Zakowski M, Rusch V, Kris M, Viale A, Motoi N, Travis W, Conley B, Seshan VE, Meyerson M, Kuick R, Dobbin KK, Lively T, Jacobson JW, Beer DG (2008) Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 14 (8): 822–827.
Song H, Hanlon N, Brown NR, Noble ME, Johnson LN, Barford D (2001) Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2. Mol Cell 7 (3): 615–626.
Welsh EA, Eschrich SA, Berglund AE, Fenstermacher DA (2013) Iterative rank-order normalization of gene expression microarray data. BMC Bioinformatics 14: 153.
Ruhe A, Wold H, Dunn WJ (1984) The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM J Sci Stat Comput 5 (3): 735–743.
Wurzenberger C, Gerlich DW (2011) Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol 12 (8): 469–482.
Yeh CT, Lu SC, Chao CH, Chao ML (2003) Abolishment of the interaction between cyclin-dependent kinase 2 and Cdk-associated protein phosphatase by a truncated KAP mutant. Biochem Biophys Res Commun 305 (2): 311–314.
Yeh CT, Lu SC, Chen TC, Peng CY, Liaw YF (2000) Aberrant transcripts of the cyclin-dependent kinase-associated protein phosphatase in hepatocellular carcinoma. Cancer Res 60 (17): 4697–4700.
Yu Y, Jiang X, Schoch BS, Carroll RS, Black PM, Johnson MD (2007) Aberrant splicing of cyclin-dependent kinase-associated protein phosphatase KAP increases proliferation and migration in glioblastoma. Cancer Res 67 (1): 130–138.
Zang X, Chen M, Zhou Y, Xiao G, Xie Y, Wang X (2015) Identifying CDKN3 gene expression as a prognostic biomarker in lung adenocarcinoma via meta-analysis. Cancer Inform 14 (Suppl 2): 183–191.
Acknowledgements
This study was supported in part by grants from Florida Biomedical Research Program (3BN01, 3KB06) and NIH (R01CA178456, P30CA076292, P50CA119997). We thank Heather Belle and Steven Enkemann for assistance in the initial analysis of protein tyrosine phosphatase expression. We have utilised publically available data from the TCGA Research Network (http://cancergenome.nih.gov/) and analysed it via cBioPortal (www.cbioportal.org). Total Cancer Care (TCC) is enabled, in part, by the generous support of the DeBartolo Family. We thank the many patients who so graciously provided data and tissues to the TCC Consortium. Our study also received valuable assistance from staffs in the Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, including Tissue, Molecular Genomics and Cancer Informatics Cores.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Fan, C., Chen, L., Huang, Q. et al. Overexpression of major CDKN3 transcripts is associated with poor survival in lung adenocarcinoma. Br J Cancer 113, 1735–1743 (2015). https://doi.org/10.1038/bjc.2015.378
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.378
- Springer Nature Limited
Keywords
This article is cited by
-
An anoikis-based gene signature for predicting prognosis in malignant pleural mesothelioma and revealing immune infiltration
Journal of Cancer Research and Clinical Oncology (2023)
-
Six genes involved in prognosis of hepatocellular carcinoma identified by Cox hazard regression
BMC Bioinformatics (2021)
-
Bioinformatics-based screening of key genes for transformation of liver cirrhosis to hepatocellular carcinoma
Journal of Translational Medicine (2020)
-
Lung squamous cell carcinoma and lung adenocarcinoma differential gene expression regulation through pathways of Notch, Hedgehog, Wnt, and ErbB signalling
Scientific Reports (2020)
-
Silencing of Synuclein-γ inhibits human cervical cancer through the AKT signaling pathway
Cellular & Molecular Biology Letters (2019)