Abstract
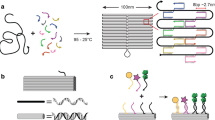
The assembly of synthetic, controllable molecular mechanical systems1,2,3,4,5,6,7 is one of the goals of nanotechnology. Protein-based molecular machines, often driven by an energy source such as ATP, are abundant in biology8,9. It has been shown previously that branched motifs of DNA can provide components for the assembly of nanoscale objects10, links11 and arrays12. Here we show that such structures can also provide the basis for dynamic assemblies: switchable molecular machines. We have constructed a supramolecular device consisting of two rigid DNA ‘double-crossover’ (DX) molecules connected by 4.5 double-helical turns. One domain of each DX molecule is attached to the connecting helix. To effect switchable motion in this assembly, we use the transition between the B and Z13,14 forms of DNA. In conditions that favour B-DNA, the two unconnected domains of the DX molecules lie on the same side of the central helix. In Z-DNA-promoting conditions, however, these domains switch to opposite sides of the helix. This relative repositioning is detected by means of fluorescence resonance energy transfer spectroscopy, which measures the relative proximity of two dye molecules attached to the free ends of the DX molecules. The switching event induces atomic displacements of 20–60 Å.


Similar content being viewed by others
References
Dado, G. & Gellman, S. H. Redox control of secondary structure in a designed peptide. J. Am. Chem. Soc. 115, 12609–12610 (1993).
Handel, T. M., Williams, S. A. & deGrado, W. F. Metal ion dependent modulation of the dynamics of a designed protein. Science 261, 879–885 (1993).
Bissell, R. A., Códova, E., Kalfer, A. E. & Stoddart, J. F. Achemically and electrochemically switchable molecular shuttle. Nature 369, 133–137 (1994).
Zelkovich, L., Libman, J. & Shanzer, A. Molecular redox switches based on chemical triggering of iron translocation in triple-stranded helical complexes. Nature 374, 790–792 (1995).
Livoreil, A. et al. Electrochemically and photochemically driven ring motions in a dissymmetrical copper [2] catenate. J. Am. Chem. Soc. 119, 12114–12124 (1997).
Murakami, M., Kawabuchi, A., Kotoo, K., Kumitake, M. & Nakashima, N. Alight-driven molecular shuttle based on a rotaxane. J. Am. Chem. Soc. 119, 7605–7606 (1997).
Zahn, S. & Canary, J. W. Redox-switched exciton-coupled circular dichroism: A novel strategy for binary molecular switching. Angew. Chem. Int. Edn 37, 305–307 (1998).
Nedelec, F. J., Surrey, T., Maggs, A. C. & Leibler, S. Self-organization of microtubules and motors. Nature 389, 305–308 (1997).
Alberts, B. The cell as a collection of protein machines. Cell 92, 291–294 (1998).
Chen, J. & Seeman, N. C. The synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Mao, C., Sun, W. & Seeman, N. C. Assembly of Borromean rings from DNA. Nature 386, 137–138 (1997).
Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
Rich, A., Nordheim, A. & Wang, A. H.-J. The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem. 53, 791–846 (1984).
Pohl, F. M. & Jovin, T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J. Mol. Biol. 67, 375–396 (1972).
Seeman, N. C. in Biomolecular Materials (ed. Viney, C., Case, S. T. & Waite, J. H.) 123–134 (Symp. Proc. Vol. 292, Materials Research Soc, (1993)).
Li, X., Yang, X., Qi, J. & Seeman, N. C. Antiparallel DNA double crossover molecules as components for nanoconstruction. J. Am. Chem. Soc. 118, 6131–6140 (1996).
Fu, T.-J. & Seeman, N. C. DNA double crossover structures. Biochemistry 32, 3211–3220 (1993).
Sheardy, R. D. et al. Sequence dependence of the free energy of B-Z junction formation in deoxyoligonucleotides. J. Mol. Biol. 231, 475–488 (1993).
Jares-Erijman, E. A. & Jovin, T. M. Determination of DNA helical handedness by fluorescence resonance energy transfer. J. Mol. Biol. 257, 597–617 (1996).
Ma, R.-I., Kallenbach, N. R., Sheardy, R. D., Petrillo, M. L. & Seeman, N. C. 3-arm nucleic acid junctions are flexible. Nucleic Acids Res. 14, 9745–9753 (1986).
Behe, M. & Felsenfeld, G. Effects of methylation on a synthetic polynucleotide: The B-Z transition in poly (dG-m5dC).poly (dG-m5dC). Proc. Natl Acad. Sci. USA 78, 1619–1623 (1981).
Seeman, N. C. Physical models for exploring DNA topology. J. Biol. Struct. Dyn. 5, 997–1004 (1988).
Seeman, N. C. DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct. 27, 225–248 (1998).
Smith, S. S. et al. Nucleoprotein-based nanoscale assembly. Proc. Natl Acad. Sci. USA 94, 2162–2167 (1997).
Yang, X., Liu, B., Vologodskii, A. V., Kemper, B. & Seeman, N. C. Torsional control of double stranded DNA branch migration. Biopolymers 45, 69–83 (1998).
Clegg, R. M. et al. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry 31, 4846–4856 (1992).
Chattopadhyaya, R., Grzeskowiak, K. & Dickerson, R. E. Structure of a T4hairpin loop on a Z-DNA stem and comparison with A-RNA and B-DNA loops. J. Mol. Biol. 211, 189–210 (1990).
Acknowledgements
We thank S. Zhang for work done on previous systems, and D. Millar, N. Geacintov and R. Sheardy for advice and for the use of equipment. This work was supported by the Office of Naval Research, the National Institute of General Medical Sciences, the National Science Foundation/DARPA and the Information Directorate of the Air Force Research Laboratory (Rome, NY).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mao, C., Sun, W., Shen, Z. et al. A nanomechanical device based on the B–Z transition of DNA. Nature 397, 144–146 (1999). https://doi.org/10.1038/16437
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/16437
- Springer Nature Limited
This article is cited by
-
Interfacing DNA nanotechnology and biomimetic photonic complexes: advances and prospects in energy and biomedicine
Journal of Nanobiotechnology (2022)
-
Self-assembled endogenous DNA nanoparticles for auto-release and expression of the eGFP gene in Bacillus subtilis
Communications Biology (2022)