Abstract
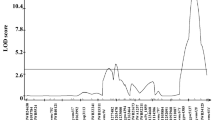
Based on different marker information content mapping of QTLs for Fusarium head blight resistance in wheat was compared with regard to number and consistency of detected QTLs as well as QTL positions and effects. Therefore, two linkage maps, obtained by ‘dominant’ and ‘codominant genotyping’ of hemizygous markers, were constructed with 211 AFLPs, 37 SSRs and the barley RGA marker XaACT/CAA. The ‘codominant marker set’ comprised 59% codominant markers, whereas the ‘dominant map’ consisted of only 13%. A segregating wheat population of 94 F4-RILs was used for QTL analysis. Fusarium head blight resistance was estimated in field trials in six environments. Conventional ‘dominant marker scoring’ found seven QTLs. The phenotypic variations explained by QTLs detected in single environment analyses ranged from 11.1 to 44.6%. QTL analysis performed with the ‘codominant marker set’ confirmed not only all QTL positions as revealed by ‘dominant QTL analysis', but also 12 additional QTLs were found. QTLs in single environments explained 36.3 up to 55.7% of the phenotypic variation. In the QTL analysis across all environments, none of the QTLs could be confirmed using ‘dominant marker scoring’. However, by ‘codominant QTL analysis' environment-specific QTLs were retrieved. STS marker XaACT/CAA was found to be significantly associated with FHB resistance only by ‘codominant scoring’. Support intervals of QTLs commonly found in both marker sets averaged to 10.3 cM in the ‘dominant QTL analysis', whereas the length was shortened to 8.9 cM by ‘codominant genotyping’. The advantages of extracting codominant information from dominant markers are discussed.
Similar content being viewed by others
References
Becker J., Vos P., Kuiper M., Salamini F. and Heun M. 1995. Combined mapping of AFLP and RFLP markers in barley. Mol. Gen. Genet. 249: 65–73.
Buerstmayr H., Lemmens M., Hartl L., Doldi L., Steiner B., Stierschneider X. and Ruckenbauer P. 2002. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (Type II resistance). Theor. Appl. Genet. 104: 84–91.
Buerstmayr H., Lemmens M., Fedak G. and Ruckenbauer P. 1999. Backcross reciprocal monosomic analysis of Fusarium head blight resistance in wheat (Triticum aestivum L.). Theor. Appl. Genet. 98: 76–85.
Castiglioni P., Ajmone-Marsan P., Van Wijk R. and Motto M. 1999. AFLP markers in a molecular linkage map of maize: Codominant scoring and linkage group distribution. Theor. Appl. Genet. 99: 425–431.
Gilbert J. and Tekauz A. 2000. Review: Recent developments in research on fusarium head blight of wheat in Canada. Can. J. Plant. Pathol. 22: 1–8.
Hallauer A.R. and Miranda J.B. 1981. Quantitative Genetics in Maize Breeding. Iowa State University Press, Ames, IA.
Hartl L., Mohler V., Zeller FJ., Hsam S.L.K. and Schweizer G. 1999. Identification of AFLP markers closely linked to the powdery mildew resistance genes Pm1c and Pm4a in common wheat (Triticum aestivum L.). Genome 42: 322–329.
Jiang C. and Zeng Z.B. 1997. Mapping quantitative trait loci with dominant and missing markers in various crosses from two in-bred lines. Genetica 101: 47–58.
Kosambi D.D. 1990. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175.
Lande R. and Thompson R. 1990. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124: 743–756.
Lander E.S. and Botstein D. 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199
Lübberstedt T., Mohler V. and Wenzel G. 2002. Function of genetic material – Genes involved in quantitative and qualitative resistance. Prog. Bot. 63: 80–105.
Maheswaran M., Subudhi P.K., Nandi S., Xu J.C., Parco A., Yang D.C. and Huang N. 1997. Polymorphism, distribution, and segregation of AFLP markers in a doubled haploid rice population. Theor. Appl. Genet. 94: 39–45.
McIntosh R.A., Hart G.E., Devos K.M., Gale M.D. and Rogers W.J. 1998. Catalogue of gene symbols for wheat: 1998 edition. http://wheat.pw.usda.gov/ggpages/wgc/98
Mohler V., Klahr A., Wenzel G. and Schwarz G. 2002. A resistance gene analog useful for targeting disease resistance genes against different pathogens on group 1S chromosomes of barley, wheat and rye. Theor. Appl. Genet. 105: 364–368.
Moreau L., Monod H., Charcosset A. and Gallais A. 1999. Marker-assisted selection with spatial analysis of unreplicated field trials. Theor. Appl. Genet. 98: 234–242.
Olson M., Hood L., Cantor C.H. and Botstein D. 1989. A common language for physical mapping of the human genome. Science 24: 1434–1435.
Roeder M.S., Plaschke J., König S.U., Boerner A., Sorrells M.E., Tanksley S.D. and Ganal M.W. 1995. Abundance, variability and chromosomal location of microsatellites in wheat. Mol. Gen. Genet. 246: 327–333.
Schwarz G., Michalek W., Mohler V., Wenzel G. and Jahoor A. 1999. Chromosome landing at the Mla locus in barley (Hordeum vulgare L.) by means of high-resolution mapping with AFLP markers. Theor. Appl. Genet. 98: 521–530.
Schwarz G., Herz M., Huang XQ., Michalek W., Jahoor A., Wenzel G. and Mohler V. 2000. Application of fluorescence-based semi-automated AFLP analysis in barley and wheat. Theor. Appl. Genet. 100: 545–551.
Spielmeyer W., Robertson M., Collins N., Leister D., Schulze-Lefert P., Seah S., Moullet O. and Lagudah E.S. 1998. A super-family of disease resistance gene analogs is located on all homoeologous chromosome groups of wheat (Triticum aestivum). Genome 41: 782–788.
Spielmeyer W. and Lagudah E.S. 2003. Homoeologous set of NBS-LRR genes located at leaf and stripe rust resistance loci on short arms of chromosome 1 of wheat. Funct. Integr. Genomics 3: 86–90.
Stam P. 1995. JoinMap 2.0 deals with all types of plant mapping populations. Plant Genome III Abstracts, http://www.intl-pag.org.
Tautz D. and Renz M. 1984. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucl. Acids. Res. 12: 4127–4138.
Toojinda T., Broers L.H., Chen X.M., Hayes P.M., Kleinhofs A., Korte J., Kudrna D., Leung H., Line R.F., Powell W., Ramsay L., Vivar H. and Waugh R. 2000. Mapping quantitative and qualitative disease resistance genes in a doubled population of barley (Hordeum vulgare). Theor. Appl. Genet. 101: 580–589.
Utz H.F. and Melchinger A.E. PLABQTL: a program for composite interval mapping of QTLs. J. Quant. Trait Loci. http://probe.nalusda.gov:8000/otherdocs/jqtl
Van Berloo R. and Stam P. 1998. Marker-assisted selection in autogamous RIL populations: a simulation study. Theor. Appl. Genet. 96: 147–157.
Van Berloo R. and Stam P. 2001. Simultaneous marker-assisted selection for multiple traits in autogamous crops. Theor. Appl. Genet. 102: 1107–1112.
Vos P., Hogers R., Bleeker M., Reijans M., Van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M. and Zabeau M. 1995. AFLP: A new technique for DNA fingerprinting. Nucl. Acids Res. 23: 4407–4414
Williams J.G.K., Kubelik A.R., Livak K.J., Rafalski J.A. and Tingey S.V. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic. Acids Res. 18: 6531–6535.
Wu R.L. 1999. Mapping quantitative trait loci by genotyping haploid tissues. Genetics 152. 1741-1752.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Klahr, A., Mohler, V., Herz, M. et al. Enhanced power of QTL detection for Fusarium head blight resistance in wheat by means of codominant scoring of hemizygous molecular markers. Molecular Breeding 13, 289–300 (2004). https://doi.org/10.1023/B:MOLB.0000034075.99093.09
Issue Date:
DOI: https://doi.org/10.1023/B:MOLB.0000034075.99093.09