Abstract
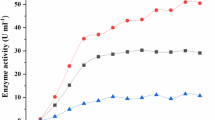
Amylomyces rouxii eliminated 85% of initial pentachlorophenol (PCP) at 12.5 mg l−1 when grown with 0.1 g tyrosine l−1, but only 55% without tyrosine. Addition of tyrosine in the culture medium increased the monophenolase activity by 1.8-fold. Tyrosinase is thus indicated to be the phenoloxidase involved in PCP degradation by A. rouxii.
Similar content being viewed by others
References
Cortés DV, Bernal R, Tomasini A (2001) Efecto de las condiciones de cultivo sumergido en la degradación de pentaclorofenol por Rhizopus nigricans. Inf. Tecnol. 12: 75-80.
Cserjesi AJ, Jhonson EL (1971) Methylation of pentachlorophenol by Trichoderma virgatum. Can. J. Microbiol. 18: 45-49.
Fernandez FJ (1997) Caracterización de la expresión de los genes implicados en la biosíntesis de penicilina: reacción enzimática catalizada por el producto del último gen (penDE) de la ruta biosintética de penicilina. Ph.D Thesis. Leon, España: Universidad de Leon.
Gukasyan GS (1999) Effect of lignin on growth and tyrosinase activity of fungi from the genus Aspergillus. Biochemistry 64: 224-227.
Mazzocco F, Pefferi PG (1976) An improvement of the spectrophotometric method for the determination of tyrosinase catecholase activity by besthor's hydrazone. Anal. Biochem. 72: 643-647.
Okeke BC, Paterson A, Smith JE, Watson-Craik IA (1997) Comparative biotransformation of pentachlorophenol in soils by solid substrate cultures of Lentinus edodes. Appl. Environ. Microbiol. 48: 563-569.
Reddy GVB, Gold MH (2000) Degradation of pentachlorophenol by Phanerochaete chrysosporium: intermediates and reactions involved. Microbiology 146: 405-413.
Sánchez-Amat A, Lucas-Elío P, Fernández E, García-Barrón JC, Solano F (2001) Molecular cloning and functional characterization of a unique multipotent polyphenol oxidase from Marinomas mediterranea. Biochim. Biophys. Acta 154: 104-116.
Seigle-Murandi F, Steiman R, Benoit-Guyod JL, Guiraud P (1993) Fungal degradation of pentachlorophenol by micromycetes. J. Biotechnol. 30: 27-35.
Smit E, Leeflang P, Glandorf B, van Elsas JD, Wernars K (1999) Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65: 2614-2621.
Tien M, Kirk TK (1988) Lignin peroxidase of Phanerochaete chrysosporium. Meth. Enzymol.: 238-249.
Tomasini A, Flores V, Cortés D, Barrios-González J (2001) An isolate of Rhizopus nigricans capable of tolerating and removing pentachlorophenol. World J. Microbiol. Biotechnol. 17: 201-205.
Tomasini-Campocosio A, Villareal-Arellanos HR, Barrios-González J (1996) Resistencia de una cepa de Rhizopus sp. al crecer en medios conteniendo pentaclorofenol. Avances Ing. Quim. 6: 36-40.
Wada S, Ichikawa H, Tatsumi K (1993) Removal of phenols from wastewater by soluble and immobilized tyrosinase. Biotechnol. Bioeng. 42: 854-858.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Montiel, A., Fernández, F., Marcial, J. et al. A fungal phenoloxidase (tyrosinase) involved in pentachlorophenol degradation. Biotechnology Letters 26, 1353–1357 (2004). https://doi.org/10.1023/B:BILE.0000045632.36401.86
Issue Date:
DOI: https://doi.org/10.1023/B:BILE.0000045632.36401.86