Abstract
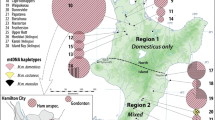
In order to contribute to knowledge of colonization patterns in the rodent Calomys musculinus, a natural reservoir of the virus producing Argentine hemorrhagic fever (AHF), we studied the haplotype diversity of the mitochondrial DNA D-loop region in five natural populations from central Argentina. Digestion with eight restriction enzymes (RsaI, MseI, Tsp509I, AluI, AciI, HaeIII, NlaIII, and AseI) revealed polymorphism in the 1300 bp fragment amplified by PCR. Twenty different composite haplotypes were detected. Hierarchical analyses indicated that almost all variation (94%) is contained within local populations. Haplotypes 1 and 2, shared by all populations, were the most frequent. Nonsignificant genetic differentiation was found among populations of the endemic and nonendemic areas of AHF. All locations sampled presented exclusive haplotypes in spite of their geographic proximity, which would support previous observations indicating restricted gene flow among C. musculinus populations.
Similar content being viewed by others
REFERENCES
Avise, J. C. (1994). Molecular Markers, Natural History and Evolution, Chapman & Hall, New York.
Avise, J. C. (2000). Phylogeography. The History and Formation of Species, Harvard University Press, Cambridge, MA.
Avise, J. C., Lansman, R. A., and Shade, R. O. (1979). The use of restriction endonucleases to measure mitochondrial sequence relatedness in natural populations. Population structure and evolution in the genus Peromyscus. Genetics 92:79.
Avise, J. C., Shapira, J. F., Daniel, S. W., Aquadro, C. F., and Lansman, R. A. (1983). Mitochondrial DNA differentiation during the speciation process in Peromyscus. Mol. Biol. Evol. 1:38.
Brown, J. R., Beckenbach, K., Beckenbach, A. T., and Smith, M. J. (1996). Length variation, heteroplasmy and sequence divergence in the mitochondrial DNA of four species of Sturgeon (Acipenser). Genetics 142:525.
Cann, R. L., Brown, W. M., and Wilson, A. C. (1984). Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics 106:479.
Casane, D., Dennebouy, N., de Rochambeau, H., Mounolou, J. C., and Monnerot, M. (1994). Genetic anlysis of systematic mitochondrial heteroplasmy in rabbits. Genetics 138:471.
Chiappero, M. B., Sabattini, M. S., Blanco, A., Calder´on, G. E., and Gardenal, C. N. (in press). Gene flowamong Calomys musculinus (Rodentia, Muridae) populations in Argentina. Genetica 114:63.
Clark, A. G. (1988). Deterministic theory of heteroplasmy. Evolution 42(3):621.
Gach, M. H., and Brown, W. M. (1997). Characteristics and distribution of large tandem duplications in Brook Stickleback (Culaea inconstans) mitochondrial DNA. Genetics 145:383.
Garc´a, J. B., Morzunov, S. P., Levis, S., Rowe, J., Calder´on, G., Enr´a, D., Sabattini, M. S., Buchmeier, M. J., Bowen, M. D., and Jeor, S. C. (2000). Genetic diversity of the Junin virus in Argentina: Geographic and temporal patterns. Virology 272:127.
Hillis, D. M., Moritz, C., and Mable, B. K. (1996). Molecular Systematics, Sinauer Ass., Massachusetts. Jaarola, M., and Tegelstr¨om, H. (1995). Colonization history of north European fields voles (Microtus agrestis) revealed by mitochondrial DNA. Mol. Ecol. 4:299.
Kessler, L. G., and Avise, J. C. (1985). Microgeographic lineage analysis by mitochondrial genotype. Variation in the cotton rat (Sigmodon hispidus). Evolution 39:831.
McElroy, D., Moran, P., Bermingham, E., and Kornfield, I (1992). REAP. An integrated environment for the manipulation and phylogenetic analysis of restriction data. J. Hered. 83:157.
Milligan, B. G. (1992). Plant DNA isolation. In Hoetzel, A. R. (ed.), Molecular Genetic Analysis of Populations, A Practical Approach, IRL Press, New York, p. 59.
Mills, J., and Childs, J. (1998). Ecologic studies of rodent reservoirs. Their relevance for human health. Emerging Infect. Dis. 4:529.
Mills, J. N., Ellis, B. A., McKee, K. T., Ksiazek, T. G., Barrera Oro, J. G., Maiztegui, J. I., Calderon, G. E., Peters, C. J., and Childs, J. E. (1991). Junin virus activity in rodents from endemic and nonedemic loci in central Argentina. Am. J. Trop. Med. Hyg. 44:589.
Nei, M., and Tajima, F. (1981). DNA polymorphism detectable by restriction endonucleases. Genetics 97:145.
Rando, J. C., Cabrera, V. M., Larruga, J. M., Hern´andez, M., Gonz´alez, A. M., Pinto, F., and Bandelt, H. J. (1999). Phylogeographic patterns of mtDNAreflecting the colonization of the Canary Islands. Ann. Hum. Genet. 63:413.
Sabattini, M. S., and Contigiani, M. S. (1982). Ecological factors influencing the maintenance of arena viruses in nature with special reference to the agents of Argentinean Hemorrhagic Fevers. International Symposium on Tropical Arenaviruses and Hemorrhagic Fevers. In de Pinheiro, F. P. (ed.), Academia Brasileira de Ciencias, Rio de Janeiro, p. 251.
Schneider, S., Kueffer, J. M., Roessli, D., and Excoffier, L. (1997). Arlequin ver 1.1. A Software for Population Genetic Data Analysis, Genetics and Biometry Laboratory, University of Geneva, Switzerland.
Smith, M. F. (1998). Phylogenetic relationships and geographic structure in pocket gophers in the genus Thomomys. Mol. Phylogenet. Evol. 9:1.
Wilkinson, G. S., Mayer, F., Kerth, G., and Petri, B. (1997). Evolution of repeated sequence arrays in the D-loop region of Bat mitochondrial DNA. Genetics 146:1035.
Xu, X., and Arnason, U. (1996). The mitochondrial DNA molecule of Sumatran orangutan and a molecular proposal for two (Bornean and Sumatran) species of orangutan. J. Mol. Evol. 43:431.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ittig, R.E.G., Gardenal, C.N. Haplotype Diversity of the Mitochondrial DNA D-Loop Region in Calomys musculinus (Rodentia, Muridae) Detected by PCR-RFLP. Biochem Genet 40, 293–302 (2002). https://doi.org/10.1023/A:1020250701882
Issue Date:
DOI: https://doi.org/10.1023/A:1020250701882