Abstract
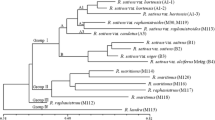
A few of the approximately 300 Cotoneaster species described are diploid but the majority appear to be polyploid. The occurrence of apomixis inpolyploid Cotoneaster species has been reported but never proven with genetic markers. We have used 76 polymorphic RAPD markers to investigate the breeding system and phenetic grouping of some critical taxa, including a total of 19 plant accessions representing 13 mostly European species in the series Cotoneaster. Three to four individual plants, raised from seed from the same original plant, were analyzed for each of three accessions to investigate the possible occurrence of apomictic seed set. Absolutely congruent RAPD profiles were encountered among seedlings from one accession, whereas we found one or two marker differences among seedlings from the other two accessions. Genetic similarities among the different accessions were analyzed with a UPGMA-derived dendrogram. The most deviant taxon was the Chinese C. albokermesinus. A group withC. soczavianus and C. tomentosus was rather isolated from the remainder, as was also C. kullensis. Among the remaining taxa, two well supported clusters were found: (1) C. antoninae and C. uralensis, and (2) C. integerrimus and C. raboutensis, whereas the other five species (C. canescens, C. niger, C. scandinavicus, C. juranus, C. cambricus) formed a poorly supported cluster with no clear substructuring. A principal coordinate analysis yielded results that were in good correspondence with the dendrogram. Again C. albokermesinus appears to be totally isolated from the other species. In addition, two well-defined and rather isolated groups were found: (1) C. tomentosus and C. soczavianus, and (2) C. antoninae and C. uralensis, with the remainder comprising a loosely defined group.
Similar content being viewed by others
References
Adams, R.P. & T. Demeke, 1993. Systematic relationships in Juniperus based on randomamplified polymorphic DNAs (RAPDs). Taxon 42: 553–571.
Asker, S.E. & L. Jerling, 1992. Apomixisin Plants. CRC Press, Boca Raton, USA.
Bartish, I., Rumpunen, K. & H. Nybom, 1999a. Genetic diversity inChaenomeles (Rosaceae) revealed by RAPD analysis. Pl Syst Evol 214: 131–145.
Bartish, I., Jeppsson, N. & H. Nybom, 1999b. Population genetic structure in the dioecious pioneer plant species Hippophae rhamnoides investigated by RAPD markers. Mol Ecol 8: 791–802.
Favarger, C., 1969. Notes de caryologie AlpineV. Bull Soc Neuchat Sci Nat 92: 13–30.
Favarger, C., 1971. Relation entre de la Flore méditerranéenne etcelle des enclaves à végétation subméditerranéenne d'Europe centrale. Boissiera 19: 149–168.
Felsenstein, J.,1993. PHYLIP (Phylogeny inference package) Version 3.5c. Department of Genetics, University of Washington, Seattle.
Flinck, K.E., Fryer, J., Garraud, L., Hylmö, B. & J. Zeller, 1998. Cotoneasterraboutensis, espèce nouvelle de l'ouest des Alpes, et révision du genre Cotoneaster dans les Alpes françaises. Bull Mens Soc Linn Lyon 67: 272–282.
Flinck, K.E. & B. Hylmö, 1962. Cotoneaster harrysmithii, a new species from WesternChina. Bot Notiser 115: 29–34.
Flinck, K.E. & B. Hylmö, 1964. Cotoneaster splendens, a new speciesfrom Western China. Bot Notiser 117: 124–132.
Flinck, K.E. & B. Hylmö, 1966. A list of series andspecies in the genus Cotoneaster. Bot Notiser 119: 445–463.
Fryer, J. & B. Hylmö, 1994. The nativeBritish Cotoneaster - Great Orme berry - renamed. Watsonia 20: 61–63.
Hjelmqvist, H., 1962. The embryosac development of some Cotoneaster species. Bot Notiser 115: 208–236.
Hylmö, B., 1993. Oxbär,Cotoneaster, i Sverige [Cotoneaster in Sweden]. Svensk Botanisk Tidskrift 87: 305–330.
Hylmö, B. & J. Fryer, 1999. Cotoneasters in Europe. Acta Bot Fennica 162: 179–184.
Krügel, T., 1992.Zur zytologischen Struktur der Gattung Cotoneaster III. Phytotaxonomie 15: 69–86.
Nei, M., 1972.Genetic distance between populations. Amer Naturalist 106: 283–292.
Nybom, H., 1996. DNAfingerprinting - A useful tool in the taxonomy of apomictic plant groups. Folia Geobot Phytotax 31: 295–304.
Page, R.D.M., 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp ApplBiosci 12: 357–358 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Phipps, J.B., Robertson, K.R., Smith, P.G. & R. Rohrer, 1990. A checklist of the subfamily Maloideae (Rosaceae). Can J Bot 68: 2209–2269.
Ploompuu, T., 1999. Eesti kodumaised tuhkpuud [Native Cotoneasters of Estonia]. Dendrological Researches in Estonia. Estonian Dendrological Society pp. 38–54.
Regel, E., 1873. Dispositio specierum et inhorto botanico imperiali petropolitano sub diu cultarum et in imperio rossico hucusque observatarum generis Cotoneaster Medicus. Acta Horti Petropolitani 2: 311–316.
Rehder, A., 1927. Manual of Cultivated Trees and Shrubs. TheMacmillan company, New York.
Sax, H. J., 1954. Polyploidy and apomixis in Cotoneaster. J Arnold Arboretum35: 334–365.
Schneller, J., Holderegger, R., Gugerli, F., Eichenberger, K. & E. Lutz, 1998. Patterns ofgenetic variation detected by RAPDs suggest a single origin with subsequent mutations and long-distance dispersal in the apomictic fern Dryopteris remota (Dryopteridaceae). Amer J Bot85: 1038–1042.
Stace, C., 1997. New Flora of theBritish Isles, 2nd ed. Press Syndicate of the University of Cambridge, Cambridge.
Ur-Rahman, H., James, D.J., Hadonou, A.M. & P.D.S. Caligari, 1997. The use of RAPD for verifying the apomictic status of seedlings of Malus species. Theor Appl Genet 95: 1080–1083.
Weising, K., Nybom, H., Wolff, K. & W. Meyer, 1994.DNA Fingerprinting in Plants and Fungi. CRC press, Boca Raton, USA.
Werlemark, G., Uggla, M. & H. Nybom,1999. Highly skewed distribution of morphological and RAPD markers in a pair of reciprocal crosses between hemisexual dogrose species, Rosa sect. Caninae. Theor Appl Genet 98: 557–563.
Williams, J.G.K., Kubelik, A.R., Livak, K.J., Rafalski, J.A. & S.V. Tingey, 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18: 6531–6535.
Zabel, H., 1898. Nachträge zur Monographie derGattung Cotoneaster in Mitteilungen den DDG 1897: 14–30. Mitt Deutsch Dendr Gesell 7: 383–385.
Zeilinga, A.E., 1964. Polyploidy in Cotoneaster. Bot Notiser 117: 262–278.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bartish, I.V., Hylmö, B. & Nybom, H. RAPD analysis of interspecific relationships in presumably apomictic Cotoneaster species. Euphytica 120, 273–280 (2001). https://doi.org/10.1023/A:1017585600386
Issue Date:
DOI: https://doi.org/10.1023/A:1017585600386