Abstract
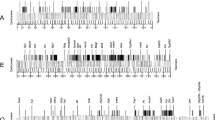
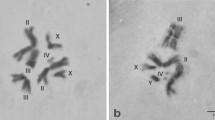
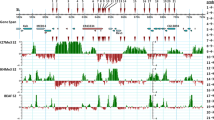
Seventeen biotin-labeled DNA sequences were hybridized to polytene chromosomes of Drosophila melanogaster and D. funebris in order to establish chromosomal homologies between these species. Ten probes correspond to cloned DNA sequences from D. melanogaster (RpII 215, MHC, H3-H4, Tor, hsp 68, hsp 28/23, hsp 83, PP1α, RpII 140, and ey), four are clones isolated from a D. subobscura genomic library (Xdh, DsubS3, DsubG3, and DsubG4), two are clones from D. funebris (F2 and Adh) and one from D. virilis (ci). The probes were chosen in order to cover all the autosomes, since X-chromosome homologies have already been studied by linkage analysis of morphological mutants. Most probes gave a unique hybridization signal; consequently, our results allow unambiguous inferences about chromosomal homologies. The results show extensive gene rearrangement within all chromosomal elements, probably due to paracentric inversions, but are consistent with Muller's proposal that chromosomal elements have conserved their genetic content during the evolution of Drosophila.
Similar content being viewed by others
References
Alonso C, Berendes HD (1975) The location of the 5S ribosomal RNA genes in Drosophila hydei. Chromosoma 51: 347-356.
Bernstein SI, Mogami K, Donady JJ, Emerson CP (1983) Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. Nature 302: 393-397.
Brock HW, Roberts DB (1983) Location of the LSP-1 genes in Drosophila species by in situ hybridization. Genetics 103: 75-92.
Casanova J, Struhl G (1989) Localized surface activity of torso, a receptor tyrosine kinase, specifies terminal body pattern in Drosophila. Genes Dev 3: 2025-2038.
Clayton FE, Guest WC (1986) Overview of chromosomal evolution in the family Drosophilidae. In: Ashburner M, Carson HL, Thompson JN, eds. The Genetics and Biology of Drosophila 3e. London: Academic Press, pp 1-38.
Cohen M Jr (1976) Ectopic pairing and evolution of 5S ribosomal RNA genes in the chromosomes of Drosophila funebris. Chromosoma 55: 349-357.
Comeron JM, Aguadé M (1996) Synonymous substitutions in the Xdh gene of Drosophila: Heterogeneous distribution along the coding region. Genetics 144: 1053-1062.
Corces V, Holmgren R, Freund R, Morimoto R, Meselson M (1980) Four heat shock proteins of Drosophila melanogaster coded within a 12-kilobase region in chromosome subdivision 67B. Proc Natl Acad Sci USA 77: 5390-5393.
Dombrádi V, Axton JM, Glover DM, Cohen PTW (1989) Cloning and chromosomal localization of Drosophila cDNA encoding the catalytic subunit of protein phosphatase 1α. High conservation between mammalian and insect sequences. Eur J Biochem 183: 603-610.
Drosopoulou E, Tsiafouli M, Mavragani-Tsipidou P, Scouras ZG (1997) The glutamate dehydrogenase, E74 and putative actin gene loci in the Drosophila montium subgroup. Chromosoma 106: 20-28.
Felger I, Pinsker W (1987) Histone gene transposition in the phylogeny of the Drosophila obscura group. Z Zool Syst Evolutforsch 25: 127-140.
Hamilton BJ, Mortin MA, Greenleaf AL (1993) Reverse genetics of Drosophila RNA polymerase II: Identification and characterization of RpII140, the genomic locus for the second-largest subunit. Genetics 134: 517-529.
Holmgren R, Livak K, Morimoto R, Freund R, Meselson M (1979) Studies of cloned sequences from four Drosophila heat shock loci. Cell 18: 1359-1370.
Jokerst RS, Weeks JR, Zerhing WA, Greenleaf AL (1989) Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet 215: 266-275
Kokoza EB, Belyaeva ES, Zhimulev IF (1992) Localization of genes esc, dor and swi in eight Drosophila species. Genetica 87: 79-85.
Lefevre G Jr (1976) A photographic representation and interpretation of the polytene chromosomes of Drosophila melanogaster salivary glands. In: Ashburner M, Novitski E, eds. The Genetics and Biology of Drosophila 1a. New York: Academic Press, pp 31-66.
Loukas M, Kafatos FC (1986) The actin loci in the genus Drosophila: Establishment of chromosomal homologies among distantly related species by in situ hybridization. Chromosoma 94: 297-308.
Matsuo Y, Yamazaki T (1989) Nucleotide variation and divergence in the histone mulitgene family in Drosophila melanogaster. Genetics 122: 87-97.
Metz CW (1916) Additional types of chromosome groups in the Drosophilidae. Am Naturalist 50: 587-599.
Montgomery E, Charlesworth B, Langley CH (1987) A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res 49: 31-41.
Muller HJ (1940) Bearings of the Drosophila work on systematics. In: Huxley J, ed. New Systematics. Oxford: Clarendon Press, pp 185-268.
Papaceit M, Juan E (1993) Chromosomal homologies between Drosophila lebanonensis and D. melanogaster determined by in situ hybridizaiton. Chromosoma 102: 361-368.
Papaceit M, Juan E (1998) Fate of dot chromosome genes in Drosophila willistoni and Scaptodrosophila lebanonensis determined by in situ hybridization. Chromosome Res 6: 49-54.
Patterson JT, Stone WS (1952) Evolution in the genus Drosophila. New York: The Macmillan Company.
Perje AM (1954) Genetic and cytological studies of Drosophila funebris. Mutations, salivary gland chromosomes and variation in sex ratios in natural and laboratory strains. Acta Zool (Stockholm) 35: 259-288.
Perje AM (1955) Genetic and cytological studies of Drosophila funebris. Some sex-linked mutations and their standard order. Acta Zool (Stockholm) 36: 51-66.
Quiring R, Walldorf U, Kloter U, Gehring WJ (1994) Homology of the eyeless gene of Drosophila to the small eye gene in mice and Aniridia in humans. Science 265: 785-789.
Ranz JM, Segarra C, Ruiz A (1997) Chromosomal homology and molecular organization of Muller's element D and E in the Drosophila repleta species group. Genetics 145: 281-295.
Segarra C, Aguadé M (1992) Molecular organization of the X chromosome in different species of the obscura group of Drosophila. Genetics 130: 513-521.
Segarra C, Lozovskaya ER, Ribó G, Aguadé M, Hartl DL (1995) P1 clones from Drosophila melanogaster as markers to study the chromosomal evolution of Muller's A element in two species of the obscura group to Drosophila. Genetics 104: 129-136.
Segarra C, Ribó G, Aguadé M (1996) Differentiation of Muller's chromosomal elements D and E in the obscura group of Drosophila. Genetics 144: 139-146.
Steinemann M (1982) Analysis of chromosomal homologies betwen two species of the subgenus Sophophora: D. miranda and D. melanogaster using cloned DNA segments. Chromosoma 87: 77-88.
Steinemann M, Pinsker W, Sperlich D (1984) Chromosome homologies within the Drosophila obscura group probed by in situ hybridization. Chromosoma 91: 46-53.
Su Y, Herrick K, Farmer JL, Jeffery DE (1992) Zaprionus tuberculatus: chromosome map and gene mapping by DNA in situ hybridization. J Hered 83: 299-304.
Tiniakov GG (1936) The inert regions and general morphology of the chromosomes in the salivary gland cells of Drosophila. Biol Zh 5: 753-802.
Tiniakov GG (1965) Salivary gland chromosome maps in Drosophila funebris. Byull Mosk Obsh Isopyt Prir Otdel Biol USSR 70: 141-144.
Tonzetich J, Hayashi S, Grigliatti TA (1990) Conservatism of sites of tRNA loci among the linkage groups of several Drosophila species. J Mol Evol 30: 182-188.
Vieira J, Vieira CP, Hartl DL, Lozovskaya ER (1997a) A framework physical map of Drosophila virilis based on P1 clones: applications in genome evolution. Chromosoma 106: 99-107.
Vieira J, Vieira CP, Hartl DL, Lozovskaya ER (1997b) Discordant rates of chromosome evolution in the Drosophila virilis species group. Genetics 147: 223-230.
Wimber DH, Wimber DR (1977) Sites of the 5S ribosomal genes in Drosophila. I. The multiple clusters in the virilis group. Genetics 86: 133-148.
Whiting JH Jr, Pliley MD, Farmer JL, Jeffery DE (1989) In situ hybridization analysis of chromosomal homologies in Drosophila melanogaster and D. virilis. Genetics 122: 99-109.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gallego, P., Juan, E. & Papaceit, M. Chromosomal Homologies Between Drosophila Melanogaster and D. Funebris Determined By In-Situ Hybridization. Chromosome Res 7, 331–339 (1999). https://doi.org/10.1023/A:1009207812569
Issue Date:
DOI: https://doi.org/10.1023/A:1009207812569