Abstract
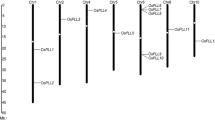
Although sequences representing members of the phytochrome (phy) family of photoreceptors have been reported in numerous species across the phylogenetic spectrum, relatively few phytochrome genes (PHY) have been fully characterized. Using rice, we have cloned and characterized the first PHYC gene from a monocot. Comparison of genomic and cDNA PHYC sequences shows that the rice PHYC gene contains three introns in the protein-coding region typical of most angiosperm PHY genes, in contrast to Arabidopsis PHYC, which lacks the third intron. Mapping of the transcription start site and 5′-untranslated region of the rice PHYC transcript indicates that it contains an unusually long, intronless, 5′-untranslated leader sequence of 715 bp. PHYC mRNA levels are relatively low compared to PHYA and PHYB mRNAs in rice seedlings, and are similar in dark- and light-treated seedlings, suggesting relatively low constitutive expression. Genomic mapping shows that the PHYA, PHYB, and PHYC genes are all located on chromosome 3 of rice, in synteny with these genes in linkage group C (sometimes referred to as linkage group A) of sorghum. Phylogenetic analysis indicates that rice phyC is closely related to sorghum phyC, but relatively strongly divergent from Arabidopsis phyC, the only full-length dicot phyC sequence available.
Similar content being viewed by others
References
Adam, E., Deak, M., Kay, S., Chua, N.H. and Nagy, F. 1993. Sequence of a tobacco (Nicotiana tabacum) gene coding for type A phytochrome. Plant Physiol. 101: 1407–1408.
Adam, E., Kozma-Bognar, L., Dallmann, G. and Nagy, F. 1995. Transcription of tobacco phytochrome-A genes initiates at multiple start sites and requires multiple cis-acting regulatory elements. Plant Mol. Biol. 29: 983–993.
Adam, E., Kozma-Bognar, L., Schäfer, E. and Nagy, F. 1997. Tobacco phytochromes: genes, structure and expression. Plant Cell Environ. 20: 678–684.
Ahn, S.N. and Tanksley, S.D. 1993. Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. USA 90: 7980–7984.
Austin, D.F. and Lee, M. 1996a. Comparative mapping in F2:3 and F6:7 generations of quantitative trait loci for grain yield and yield components in maize. Theor. Appl. Genet. 92: 817–826.
Austin, D.F. and Lee, M. 1996b. Genetic resolution and verification of quantitative trait loci for flowering and plant height with recombinant inbred lines of maize. Genome 39: 957–968.
Causse, M., Fulton, T.M., Cho, Y.G., Ahn, S.N., Chunwongse, J., Wu, K., Xiao, J., Yu, Z., Ronald, P.C., Harrington, S.E., Second, G.A., McCouch, S.R. and Tanksley, S.D. 1994. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251–1274.
Chao, S., Baysdorfer, C., Heredia-Diaz, O., Musket, T., Xu, G. and Coe, E.H. 1994. RFLP mapping of partially sequenced leaf cDNA clones in maize. Theor. Appl. genet. 88: 717–721.
Childs, K.L., Miller, F.R., Cordonnier-Pratt, M.-M., Pratt, L.H., Morgan, P.W. and Mullet, J.E. 1997. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 113: 611–619.
Christensen, A.H. and Quail, P.H. 1989. Structure and expression of a maize phytochrome-encoding gene. Gene 85: 381–390.
Church, G.M. and Gilbert, W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995.
Clapham, D.H., Kolukisaoglu, H.Ñ., Larsson, C.-T., Qamaruddin, M., Ekberg, I., Wiegmann-Eirund, C., Schneider-Poetsch, H.A.W. and von Arnold, S. 1999. Phytochrome types in Picea and Pinus. Expression patterns of PHYA-related types. Plant Mol. Biol. 40: 669–678.
Cowl, J.S., Hartley, N., Xie, D.-X., Whitelam, G.C., Murphy, G.P. and Harberd, N.P. 1994. The PHYC gene of Arabidopsis. Absence of the third intron found in PHYA and PHYB. Plant Physiol. 106: 813–814.
Davison, A.J. and Moss, B. 1989. Structure of vaccinia virus early promoters. J. Mol. Biol. 210: 749–769.
Dehesh, K., Franci, C., Sharrock, R.A., Somers, D.E., Welsch, J.A. and Quail, P.H. 1994. The Arabidopsis phytochrome A gene has multiple transcription start sites and a promoter sequence motif homologous to the repressor element of monocot phytochrome A genes. Photochem. Photobiol. 59: 379–384.
Dehesh, K., Tepperman, J., Christensen, A.H. and Quail, P.H. 1991. phyB is evolutionarily conserved and constitutively expressed in rice-seedling shoots. Mol. Gen. Genet. 225: 305–313.
Devlin, P.F., Patel, S.R. and Whitelam, G.C. 1998. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479–1487.
Devlin, P.F., Rood, S.B., Somers, D.E., Quail, P.H. and Whitelam, G.C. 1992. Photophysiology of the elongated internode (ein) mutant of Brassica rapa: the ein mutant lacks a detectable phytochrome B-like polypeptide. Plant Physiol. 100: 1442–1447.
Geballe, A.P. 1996. Translational control mediated by upstream AUG codons. In: J.W.B. Hershey, M.B. Mathews and N. Sonenberg (Eds.) Translational Control, Cold Spring Harbor Laboratory Press, Plainview, NY, p. 173.
Goosey, L., Palecanda, L. and Sharrock, R.A. 1997. Differential patterns of expression of the Arabidopsis PHYB, PHYD, and PHYE phytochrome genes. Plant Physiol. 115: 959–969.
Halliday, K.J., Thomas, B. and Whitelam, G.C. 1997. Expression of heterologous phytochromes A, B, or C in transgenic tobacco plants alters vegetative development and flowering time. Plant J. 12: 1079–1090.
Hanelt, S., Braun, B., Marx, S. and Schneider-Poetsch, H. 1992. Phytochrome evolution: a phylogentic tree with the first complete sequence of phytochrome A of a cryptogamic plant (Selaginella martensii Spring). Photochem. Photobiol. 56: 751–758.
Hershey, H.P., Barker, R.F., Idler, K.B., Lissemore, J.L. and Quail, P.H. 1985. Analysis of cloned cDNA and genomic sequences for phytochrome: complete amino acid sequence for two gene products expressed in etiolated Avena. Nucl. Acids Res. 13: 8543–8559.
Hershey, H.P., Barker, R.F., Idler, K.B., Murray, M.G. and Quail, P.H. 1987. Nucleotide sequence and characterization of a gene encoding the phytochrome polypeptide from Avena. Gene 61: 339–348.
Heun, M., Kennedy, A.E., Anderson, J.A., Lapitan, N.L.V., Sorrells, M. et al. 1991. Construction of restriction fragment length polymorphism map for barley (Hordeum vulgare). Genome 34: 437–447.
Heyer, A. and Gatz, C. 1992a. Isolation and characterization of a cDNA-clone coding for potato type A phytochrome. Plant Mol. Biol. 18: 535–544.
Heyer, A. and Gatz, C. 1992b. Isolation and characterization of a cDNA-clone coding for potato type B phytochrome. Plant Mol. Biol. 20: 589–600.
Hughes, J., Lamparter, T. and Mittmann, F. 1996. CpPHY2 (PHYCER2), a ‘normal’ phytochrome in Ceratodon. Plant Physiol. 112: 446.
Kaneko, T., Matsubayashi, T., Sugita, M. and Suguira, M. 1996. Physical and gene maps of the unicellular cyanobacterium Synechococcus sp. strain PCC6301 genome. Plant Mol. Biol. 31: 193–201.
Kay, S.A., Keith, B., Shinozaki, K., Chye, M.-L. and Chua, N.-H. 1989a. The rice phytochrome gene: structure, autoregulated expression, and binding of GT-1 to a conserved site in the 5′ upstream region. Plant Cell 1: 351–360.
Kay, S.A., Keith, B., Shinozaki, K. and Chua, N.-H. 1989b. The sequence of the rice phytochrome gene. Nucl. Acids Res. 17: 2865–2866.
Kendrick, R.E., Kerchoffs, L.H.J., van Tuinen, A. and Koornneef, M. 1997. Photomorphogenic mutants of tomato. Plant Cell Environ. 20: 746–751.
Kendrick, R.E. and Kronenberg, G.H.M. 1994. Photomorphogenesis in Plants, 2nd ed., Kluwer Academic Publishers, Dordrecht, Netherlands.
Kosambi, D.D. 1944. The estimation of map distances from recombination values. Ann. Genet. 12: 172–175.
Lagarias, D.M., Wu, S.-H., Lagarias, J.C. 1995. Atypical phytochrome gene structure in the green alga Mesotaenium caldariorum. Plant Mol. Biol. 29: 1127–1142.
Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M., Lincoln, S.E. and Newburg, L. 1987. MAPMAKER: an interactive computer package for maps of experimental and natural populations. Genomics 1: 174–181.
Lazarova, G.I., Kerckhoffs, L.H.J., Brandstädter, J., Matsui, M., Kendrick, R.E., Cordonnier-Pratt, M.-M. and Pratt, L.H. 1998a. Molecular analysis of a PHYA in wild-type and phytochrome Adeficient mutants of tomato. Plant J. 14: 653–662.
Lazarova, G.I., Kubota, T., Frances, S., Peters, J.L., Hughes, M.J.G., Brandstädter, J., Széll, M., Matsui, M., Kendrick, R.E., Cordonnier-Pratt, M.-M. and Pratt, L.H. 1998b. Characterization of tomato PHYB1 and identification of molecular defects in four mutant alleles. Plant Mol. Biol. 38: 1137–1146.
Ló pez-Juez, E., Nagatani, A., Tomizawa, K.-I., Deak, M., Kern, R., Kendrick, R.E. and Furuya, M. 1992. The cucumber long hypocotyl mutant lacks a light-stable phyB-like phytochrome. Plant Cell 4: 241–251.
Mathews, S. and Sharrock, R.A. 1996. The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol. Biol. Evol. 13: 1141–1150.
Mathews, S. and Sharrock, R.A. 1997. Phytochrome gene diversity. Plant Cell Environ. 20: 666–671.
Mathews, S., Lavin, M. and Sharrock, R.A. 1995. Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Ann. Miss. Bot. Garden 82: 296–321.
Matz, M., Shagin, D., Bogdanova, E., Britanova, O., Lukyanov, S., Diatchenko, L. and Chenchik, A. 1999. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucl. Acids Res. 27: 1558–1560.
Maucher, P.H. 1994. Molekularbiologie der Phytochrome des Farns Anemia phyllitidis (L.) SW. Thesis, University of Ulm, Ulm, Germany.
McCouch, S.R., Kochert, G., Yu, Z.H., Wang, Y.Z., Khush, G.S., Coffman, R. and Tanksley, S.D. 1988. Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76: 815–829.
Ming, R., Liu, S.-C., Lin, Y.-R., da Silva, J., Wilson, W., Braga, D., van Deyzne, A., Wenslaff, T.F., Wu, K.K., Moore, P.H., Burnquist, W., Sorrells, M.E., Irvine, J.E. and Paterson, A.H. 1998. Detailed alignment of saccharum and sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics 150: 1663–1682.
O'Donoughue, L.S., Wang, Z., Roder, M., Kneen, B., Legget, M. et al. 1992. An RFLP-based map of oats on a cross between two diploid taxa (Avena atlantica x A. hirtula). Genome 35: 765–771.
Oh, S.-K., Scott, M.P. and Sarnow, P. 1992. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 6: 1643–1653.
Paterson, A.H., Lin, Y.-R., Li, Z., Schertz, K.F., Doebley, J.F., Pinson, S.R.M., Liu, S.-C., Stansel, J.W. and Irvine, J.E. 1995. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269: 1714–1717.
Pereira, M.G., Lee, M., Bramel-Cox, P., Woodman, W., Doebley, J. and Whitkus, R. 1994. Construction of an RFLP map in sorghum and comparative mapping in maize. Genome 37: 236–243.
Pratt, L.H., Cordonnier-Pratt, M.-M., Kelmenson, P.M., Lazarova, G.I., Kubota, T. and Alba, R.M. 1997. The phytochrome gene family in tomato (Solanum lycopersicum L.). Plant Cell Environ. 20: 672–677.
Qin, M., Kuhn, R. and Quail, P.H. 1997. Overexpressed phytochrome C has similar photosensory specificity to phytochrome B but a distinctive capacity to enhance primary leaf expansion. Plant J. 12: 1163–1172.
Quail, P.H. 1994a. Photosensory perception and signal transduction in plants. Curr. Opin. Genet. Dev. 4: 652–661.
Quail, P.H. 1994b. Phytochrome genes and their expression. In: R.E. Kendrick and G.H.M. Kronenberg (Eds.) Photomorphogenesis in Plants, 2nd ed., Kluwer Academic Publishers, Dordrecht, Netherlands, pp. 71–104.
Quail, P.H. 1997a. An emerging molecular map of the phytochromes. Plant Cell Environ. 20: 657–665.
Quail, P.H. 1997b. The phytochromes: a biochemical mechanism of signaling in site? BioEssays 19: 571–579.
Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y. and Wagner, D. 1995. Phytochromes: photosensory perception and signal transduction. Science 268: 675–680.
Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M. and Chory, J. 1993. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157.
Sato, N. 1988. Nucleotide sequence and expression of the phytochrome gene in Pisum sativum: differential regulation by light of multiple transcripts. Plant Mol. Biol. 11: 697–710.
Schneider-Poetsch, H.A.W., Kolukisaoglu, Ñ., Clapham, D.H., Hughes, J. and Lamparter, T. 1998. Non-angiosperm phytochromes and the evolution of vascular plants. Physiol. Plant. 102: 612–622.
Schwer, B., Mao, X. and Shuman, S. 1998. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucl. Acids Res. 26: 2050–2057.
Senior, M.L., Chin, E.C.L., Lee, M., Smith, J.S.C. and Stuber, C.W. 1996. Simple sequence repeat markers developed from maize sequences found in the GenBank database: map construction. Crop Sci. 36: 1676–1683.
Sharrock, R.A. and Quail, P.H. 1989. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3: 1745–1757.
Shen, B., Carneiro, N., Torres-Jerez, I., Stevenson, B., McCreery, T., Helentjaris, T., Baysdorfer, C., Almira, E., Ferl, R.J., Habben, J. and Larkins, B.A. 1994. Partial sequencing and mapping of clones from two maize cDNA libraries. Plant Mol. Biol. 26: 1085–1101.
Shima, D.T., Kuroki, M., Deutsch, U., Ng, Y.-S., Adamis, A.P. and D'Amore, P.A. 1996. The mouse gene for vascular endothelial growth factor. J. Biol. Chem. 271: 3877–3883.
Singh, K., Ishii, T., Parco, A., Huang, N., Brara, D.S. and Khush, G.S. 1996. Centromere mapping and orientation of the molecular linkage map of rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 93: 6163–6168.
Somers, D.E. and Quail, P.H. 1995. Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 7: 413–427.
Tahir, M., Kanegae, H. and Takano, M. 1998. PHYC (Phytochrome C) gene in rice: isolation and characterization of a complete coding sequence. Plant Physiol. 118: 1535.
Wada, M., Kanegae, T., Nozue, K. and Fukuda, S. 1997. Cryptogram phytochromes. Plant Cell Environ. 20: 685–690.
Whitelam, G.C. and Devlin, P.F. 1997. Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 20: 752–758.
Wilson, W.A., Harrington, S.E., Woodman, W.L., Lee, M., Sorrells, M.E. and McCouch, S.R. 1999. Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize, and the domesticated panicoids. Genetics 153: 453–473.
Wu, S.-H. and Lagarias, J.C. 1997. The phytochrome photoreceptor in the green alga Mesotaenium caldariorum: implication for a conserved mechanism of phytochrome action. Plant Cell Environ. 20: 691–699.
Yeh, K.-C. and Lagarias, J.C. 1998. Eukaryotic phytochromes: light-regulated serine/theonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95: 13976–13981.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Basu, D., Dehesh, K., Schneider-Poetsch, HJ. et al. Rice PHYC gene: structure, expression, map position and evolution. Plant Mol Biol 44, 27–42 (2000). https://doi.org/10.1023/A:1006488119301
Issue Date:
DOI: https://doi.org/10.1023/A:1006488119301