Abstract
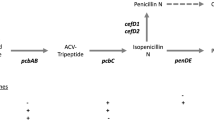
The genes pcbAB, pcbC and penDE encoding enzymes that catalyze the three steps of the penicillin biosynthesis have been cloned from Penicillium chrysogenum and Aspergillus nidulans. They are located in a cluster in Penicillium chrysogenum, Penicillium notatum, Aspergillus nidulans and Penicillium nalgiovense. The three genes are clustered in chromosome I (10.4 Mb) of P. chrysogenum, in chromosome II of P. notatum (9.6 Mb) and in chromosome VI (3.0 Mb) of A. nidulans. The cluster of the penicillin biosynthetic genes is amplified in strains with high level of antibiotic production. About five to six copies of the cluster are present in the AS-P-78 strain and 11 to 14 copies in the E1 strain (an industrial isolate), whereas only one copy is present in the wild type (NRRL 1951) strain and in the low producer Wis 54-1255 strain. The amplified region in strains AS-P-78 and E1 is arranged in tandem repeats of 106.5 or 57.6-kb units, respectively. In Acremonium chrysogenum the genes involved in cephalosporin biosynthesis are separated in at least two clusters. The pcbAB and pcbC genes are linked in the so-called ‘early cluster’ of genes involved in the cephalosporin biosynthesis. The ‘late cluster’, which includes the cefEF and cefG genes, is involved in the last steps of cephalosporin biosynthesis. The ‘early cluster’ was located in chromosome VII (4.6 Mb) in the C10 strain and the ‘late cluster’ in chromosome I (2.2 Mb). Both clusters are present in a single copy in the A. chrysogenum genome, in the wild-type and in the high cephalosporin-producing C10 strains.
Similar content being viewed by others
References
Aharanowitz Y, Cohen G & Martín JF (1992) Penicillin and cephalosporin biosynthetic genes: Structure, organization, regulation, and evolution. Annu Rev Microbiol 46: 461–495
Alvarez E, Cantoral JM, Barredo JL, Díez B & Martín JF (1987) Purification to homogeneity and characterization of acyl coenzyme A:6-aminopenicillanic acid acyltransferase of Penicillium chrysogenum. Antimicrob Agents Chemother 31: 1675–1682
Aramayo R, Adams TH & Timberlake WE (1989) A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics 122: 65–71
Baldwin JE, Adlington RM, Coates B, Crabbe MJC, Crouch NP, Keeping JW, Knight GC, Schofield CJ, Ting H-H, Vallejo CA, Thorniley M & Abraham EP (1987) Purification and initial characterization of an enzyme with deacetoxycephalosporin C synthetase and hydroxylase activities. Biochem J 245: 831–841
Baldwin JE, Bird JW & Field RA (1990) Isolaion and partial characterisation of ACV synthetase from Cephalosporium acremonium and Streptomyces clavuligerus. J Antibiot 43: 1055–1057
Barredo JL, Cantoral JM, Alvarez E, Díez B & Martín JF (1989a) Cloning, sequence analysis and transcriptional study of the isopenicillin N synthase of Penicillium chrysogenum AS-P-78. Mol Gen Genet 216: 91–98
Barredo JL, van Solingen P, Díez B, Alvarez E, Cantoral JM, Kattevilder A, Smaal EB, Groenen MAM, Veenstra AE & Martín JF (1989b) Cloning and characterization of acyl-CoA:6-APA acyl-transferase gene of Penicillium chrysogenum. Gene 83: 291–300
Barredo JL, Díez B, Alvarez E & Martín JF (1989c) Large amplification of a 35 kb DNA fragment carrying two penicillin biosynthetic genes in high penicillin producing strains of Penicillium chrysogenum. Curr Genet 16: 453–459
Bidenne C, Blondin B, Dequin S & Vezinhet F (1992) Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr Genet 22: 1–7
Brody H & Carbon J (1989) Eletrophoretic karyotype of Aspergillus nidulans. Proc Natl Acad Sci USA 86: 6260–6263
Cantoral JM, Gutiérrez S, Fierro F, Gil-Espinosa S, van Liempt H & Martín JF (1993) Biochemical characterization and molecular genetics of nine mutants of Penicillium chrysogenum impaired in penicillin biosynthesis. J Biol Chem 268: 737–744
Carr LG, Skatrud PL, Scheetz ME II, Queener SW & Ingolia TD (1986) Cloning and expression of the isopenicillin N synthetase gene from Penicillium chrysogenum. Gene 48: 257–266
Carter GL, Allison D, Rey MW & Dunn-Coleman NS (1992) Chromosomal and genetic analysis of the electrophoretic karyotype of Trichoderma reesei: mapping of the cellulase and xylanase genes. Mol Microbiol 6: 2167–2174
Chu G, Vollrath D & Davis RW (1986) Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234: 1582–1585
Clutterbuck AJ (1987) Genetic Maps. In: O'Brien SJ (ED) (pp 325–335). Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Cooley RN & Caten CE (1991) Variation in electrophoretic karyotype between strains of Septoria nodorum. Mol Gen Genet 228: 17–23
Dawkins HJS (1989) Large DNA separation using field alternation agar gel electrophoresis. J Chromatogr 492: 615–639
Demain AL (1983) Strain exchange between industry and academia. ASM News 49: 431
Díez B, Barredo JL, Alvarez E, Cantoral JM, van Solingen P, Groenen MAM, Veenstra AE & Martín JF (1989) Two genes involved in penicillin biosynthesis are linked in a 5.1 kb fragment in the genome of Penicillium chrysogenum. Mol Gen Genet 218: 572–576
Díez B, Gutiérrez S, Barredo J, van Solingen P, van der Vort LHM & Martín JF (1990) The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the α-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem 265: 16358–16365
Doolittle RF, Feng DF, Johnson MS & McClure MA (1986) Relationship of human protein sequences to those of other organisms. Symp Quant Biol 51: 447–455
Doolittle RF, Anderson KL & Feng D-F (1989) Estimating the prokaryote-eukaryote divergence time from protein sequences. In: Fernholm B, Bremer H, Jornvall H (Ed) The Hierarchy of Life (pp 73–85). Elsevier, Amsterdam
Dotzlaf JE & Yeh W-K (1987) Copurification and characterization of deacetoxycephalosporin C synthetase/hydroxylase from Cephalosporium acremonium. J Bacteriol 169: 1611–1618
Fawcett PA, Usher JJ, Huddleston JA, Bleaney RC, Nisbet JJ & Abraham EP (1976) Synthesis of δ-(L-α-aminoadipyl)cysteinyl valine and its role in penicillin biosynthesis. Biochem J 157: 651–660
Fernández FJ, Gutiérrez S, Velasco J, Montenegro E, Marcos AT & Martín JF (1994) Molecular characterization of three loss-of-function mutations in the isopenicillin N-acyltransferase gene (penDE) of Penicillium chrysogenum. J Bacteriol 176: 4941–4948
Fierro F, Gutiérrez S, Díez B & Martín JF (1993) Resolution of four large chromosomes in penicillin-producing filamentous fungi: the penicillin gene cluster is located on chromosome II (9.6 Mb) in Penicillium notatum and chromosome I (10.4 Mb) in Penicillium chrysogenum. Mol Gen Genet 241: 573–578
Fierro F, Barredo JL, Díez B, Gutiérrez S, Fernández FJ & Martín JF (1995) The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences. Proc Natl Acad Sci USA 92: 6200–6204
Fierro F, Montenegro E, Gutiérrez S & Martín JF (1996) Mutants blocked in penicillin biosynthesis show a deletion of the entire penicillin gene cluster at a specific site within a conserved hexanucleotide sequence. Appl Microbiol Biotechnol 44: 597–604
Fujisawa Y & Kanzaku T (1975) Role of acetyl-CoA:deacetylcephalosporin C acetyltransferase in cephalosporin C biosynthesis by Cephalosporium acremonium. Agric Biol Chem 39: 2043–2048
Glass NL & Kuldau GA (1992) Mating type and vegetative incompatibility in filamentous ascomycetes. Annu Rev Phytopathol 30: 201–224
Gutiérrez S, Díez B, Alvarez E, Barredo JL & Martín JF (1991a) Expression of the penDE gene of Penicillium chrysogenum encoding isopenicillin N-acyltransferase in Cephalosporium acremonium: production of benzylpenicillin by the transformants. Mol Gen Genet 225: 56–64
Gutiérrez S, Díez B, Montenegro E & Martín JF (1991b) Characterization of the Cephalosporium acremonium pcbAB gene encoding α-aminoadipyl-cysteinyl-valine synthetase, a large multidomain peptide synthetase: Linkage to the pcbC gene as a cluster of early cephalosporin biosynthetic genes and evidence of multiple functional domains. J Bacteriol 173: 2354–2365
Gutiérrez S, Velasco J, Fernánder FJ & Martín JF (1992) The cefG gene of Cephalosporium acremonium is linked to the cefEF gene and encodes a deacetylcephalosporin C acetyltransferase closely related to homoserine O-acetyltransferase. J Bacteriol 174: 3056–3064
Heinemann JA & Sprague GF (1989) Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340: 205–209
Hensel R, Zwickl P, Fabry S, Lang J & Palm P (1989) Sequence comparison of glyceraldehyde-3-phosphate dehydrogenase from three unkingdoms: evolutionary implications. Can J Microbiol. 35: 81–85
Holt G, Edwards GF & MacDonald KD (1976) The genetic of mutants impaired in the biosynthesis of penicillin. In: MacDonald KD (Ed) Second International Symposium on the Genetics of Industrial Microorganisms (pp 199–211). Academic Press, London
Jayatilake GS, Huddleston JA & Abraham EP (1982) Conversion of isopenicillin N into penicillin N in cell-free extracts of Cephalosporium acremonium. Curr Genet. 18: 523–530
Käfer E (1958) An eight-chromosome map of Aspergillus nidulans. Adv Genet 9: 105–145
Keller NP & Hohn TM (1997) Metabolic pathway gene clusters in filamentous fungi. Fungal Genet Biol 21: 17–29
Kleinkauf H & von Döhren H (1990) Nonribosomal biosynthesis of peptide antibiotics. Eur J Biochem 192: 1–15
Kleinkauf H & von Döhren H (1996) A nonribosomal system of peptide biosynthesis. Eur J Biochem 236: 335–351
Konomi T, Herchen S, Baldwin JE, Yoshida M, Hunt NA & Demain AL (1979) Cell-free conversion of δ-(L-α-aminoadipyl)-L-cysteinyl-D-valine into an antibiotic with the properties of isopenicillin N in Cephalosporium acremonium. Biochem J 184:427–430
Kupka J, Shen YQ, Wolfe S & Demain AL (1983) Partial purification and properties of the α-ketoglutarate-linked ring-expansion enzyme of β-lactam biosynthesis of Cephalosporium acremonium. FEMS Microbiol Lett 16: 1–6
Landan G, Cohen F, Aharonowitz Y, Shuali Y, Graur D & Shiffman D (1990) Evolution of isopenicillin N synthase genes may have involved horizontal gene transfer. Mol Biol Evol 7:399–406
Lein J (1986) The Panlabs penicillin strain improvement program. In: Vanek Z, Hostalek Z (Ed) Overproduction of microbial metabolites (pp 105–139). Butterworths, Boston
Leslie JF (1993) Fungal vegetative incompatibility. Annu Rev Phytopathol 31: 127–150
MacCabe AP, Riach MBR, Unkles SE & Kinghorn JR (1990) The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J 9: 279–287
MacCabe AP, van Liempt H, Palissa H, Unkles SE, Riach MBR, Pfeifer E, von Döhren H & Kinghorn JR (1991) δ-(L-α-Aminoadipyl)-L-cysteinyl-D-valine synthetase from Aspergillus nidulans: molecular characterization of the acvA gene encoding the first enzyme of the penicillin biosynthetic pathway. J Biol Chem 266: 12646–12654
Macdonald KD (1983) Fungal genetics and antibiotic production. In Vining LC (Ed) Biochemistry and genetic regulation of commercially important antibiotics. Addison-Wesley, Reading, MA: 25
Magee PT (1993) Variations in chromosome size and organization in Candida albicans and Candida stellatoidea. Trends Microbiol 1: 338–342
Makins JF, Holt G & MacDonald KD (1983) The genetic location of three mutations impairing penicillin production in Aspergillus nidulans. J Gen Microbiol 129: 3027–3033
Martín JF & Gutiérrez S (1992) Molecular genetics of fungal secondary metabolites. In: Kinghorn JR, Turner G (Ed) Applied Molecular Genetics of Filamentous Fungi (pp 214–252). Blackie and Son Ltd., Glasgow
Martín JF & Gutiérrez S (1995) Genes for β-lactam antibiotic biosynthesis. Antonie van Leeuwenhoek 67: 181–200
Martín JF, Gutiérrez S & Demain AL (1997) β-Lactams. In: Anke T (Ed) Fungal Biotechnology (pp 91–117). Chapman & Hall GmbH, Weinheim, Germany
Mathison L, Soliday C, Stepan T, Aldrich T & Rambosek J (1993).Cloning, characterization, and use in strain improvement of the Cephalosporium acremonium gene cefG encoding acetyltransferase. Curr Genet 23: 33–41
Matsuda A, Sugiura H, Matsuyama K, Matsumoto H, Ichikawa S & Komatsu K-I (1992) Molecular cloning of acetyl coenzyme A:deacetylcephalosporinCO-acetyltransferase cDNA from Acremonium chrysogenum: sequence and expression of catalytic activity in yeast. Biochem Biophys Res Commun 182: 995–1001
Montenegro E, Barredo JL, Gutiérrez S, Díez B, Alvarez E & Martín JF (1990) Cloning, characterization of the acyl-CoA:6-aminopenicillanic acid acyltransferase gene of Aspergillus nidulans and linkage to the isopenicillin N synthase gene. Mol Gen Genet 221: 322–330
Montenegro E, Fierro F, Fernández FJ, Gutiérrez S & Martín JF (1992) Resolution of chromosomes III and VI of Aspergillus nidulans by pulsed-field gel electrophoresis shows that the penicillin biosynthetic pathway genes pcbAB, pcbC and penDE are clustered on chromosome VI (3.0 Mb). J Bacteriol 174: 7063–7067
Müller WH, van de Krift TP, Kronwer AJJ, Wösten HAB, van der Voort LHM, Smaal EB & Verkleij AJ (1991) Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J 10: 489–495
Ness F & Aigle M (1995) RTM1: a member of a new family of telomeric repeated genes in yeast. Genetics 140: 945–956
Normansell PJM, Normansell ID & Holt G (1979) Genetic and biochemical studies of mutants of Penicillium chrysogenum impaired in penicillin production. J Gen Microbiol 112: 113–126
Peñalva MA, Vian A, Patiño C, Pérez Aranda A & Ramón D (1989) Molecular biology of penicillin production in Aspergillus nidulans. In: Hershberger CL, Queener SV, Hegeman G (Ed) Genetics and Molecular Biology of Industrial Microorganisms (pp 256–262). ASM, Washington, DC
Peñalva MA, Moya A, Dopazo J & Ramón D (1990) Sequences of isopenicillin N synthetase genes suggest horizontal gene transfer from prokaryotes to eukaryotes. Proc R Soc London 241: 164–169
Petes TD, Malone RE & Symington LS (1991) Recombination in yeast. In Broach JR, Pringle JR, Jones EW (Ed) The Molecular and Celular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics, Vol. I (pp 407–521). Cold Spring Harbor Laboratory Press, New York
Ramón D, Carramolin L, Patino C, Sanchez F & Peñalva MA (1987) Cloning and characterization of the isopenicillin N synthetase gene mediating the formation of the β-lactam ring in Aspergillus nidulans. Gene 57: 171–181
Ramos FR, López-Nieto MJ & Martín JF (1985) Isopenicillin N synthetase of Penicillium chrysogenum, an enzyme that converts δ-(L-α-aminoadipyl)-L-cysteinyl-D-valine to isopenicillin N. Antimicrob Agents Chemother 27: 380–387
Ramos FR, López-Nieto MJ & Martín JF (1986) Coordinate increase of isopenicillin N synthetase, isopenicillin N epimerase and deacetoxycephalosporin C synthetase in a high cephalosporin-producing mutant of Acremonium chrysogenum and simultaneous loss of the three enzymes in a non-producing mutant. FEMS Microbiol Lett 35: 123–127
Rank GH, Casey GP, Xiao W & Pringle AT (1991) Polymorphism within the nuclear and 2 μm genomes of Saccharomyces cerevisiae. Curr Genet 20: 189–194
Roach PL, Clifton IJ, Fulop V, Harlos K, Barton GJ, Hajdu J, Andersson I, Schofield CJ & Baldwin JE (1995) Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature 375: 700–704
Rustchenko EP, Curran TM & Sherman F (1993) Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol 175: 7189–7199
Samson SM, Belagaje R, Blankenship DT, Chapman JL, Perry D, Skatrud PL, Frank RM, Abraham EP, Baldwin JE, Queener SE & Ingolia TD (1985) Isolation, sequence determination and expression in E. coli of the isopenicillin N synthetase gene from Cephalosporium acremonium. Nature 318: 191–194
Samson SM, Dotzlaf JE, Slisz ML, Becker GW, van Frank RM, Veal LE, Yeh W-K, Miller JE, Queener SW & Ingolia TD (1987) Cloning and expression of the fungal expandase/hydroxylase gene involved in cephalosporin biosynthesis. Bio/Technology 5: 1207–1214
Scheidegger A, Kuenzi MT & Nuesch J (1984) Partial purification and catalytic properties of a bifunctional enzyme in he biosynthetic pathway of β-lactams in Acremonium chrysogenum. J Antibiot 37: 522–531
Schwartz DC & Cantor CR (1984) Separation of yeast chromosome-sized DNA by pulse field gradient gel electrophoresis. Cell 37: 67–75
Selker EU (1990) Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet 24: 579–613
Sermonti G (1956) Complementary genes which affect penicillin yields. J Gen Microbiol 15: 599–608
Skatrud PL & Queener SW (1989) An electrophoretic molecular karyotype for an industrial strain of Cephalosporium acremonium. Gene 78: 331–338
Skatrud PL, Tietz AJ, Ingolia TD, Cantwell CA, Fisher DL, Chapman JL & Queener SW (1989) Use of recombinant DNA to improve production of cephalosporin C by Cephalosporium acremonium. Bio/Technology 7: 477–485
Smith DJ, Burnham MKR, Edwards J, Earl AJ & Turner G (1990a) Cloning and heterologous expression of the penicillin biosynthetic gene cluster form Penicillium chrysogenum. Bio/Technology 8: 39–41
Smith DJ, Burnham MKR, Bull JH, Hodgson JE, Ward JM, Browne P, Brown J, Barton B, Earl AJ & Turner G (1990b) β-Lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J 9: 741–747
Smith AW, Collins K, Ramsden M, Fox HM & Peberdy J (1991) Chromosome rearrangements in improved cephalosporin C-producing strains of Acremonium chrysogenum. Curr Genet 19: 235–237
Smith MW, Feng D & Doolittle RF (1992) Evolution by acquisition: The case for horizontal gene transfers. Trends Biochem Sci 17: 489–493
Talbot NJ, Salch YP, Ma M & Hamer JE (1993) Karyotypic variation within clonal lineages of the rice blast fungus, Magnaporthe grisea. Appl Environ Microbiol 59: 585–593
Turgay K, Krause M & Marahiel MA (1992) Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate forming enzymes. Mol Microbiol 6: 529–546
Vollrath D & Davis RW (1987) Resolution of DNA molecules greater than 5 mega bases by contour-clamped homogeneous electric fields. Nucleic Acids Res 15: 7865–7876
Walton JD, Paquin CE, Kaneko K & Williamson VM (1986) Resistance to antimycin A in yeast by amplification of ADH4 on a linear, 42 kb palindromic plasmid. Cell 46: 857–863
Walz M & Kück U (1991) Polymorphic karyotypes in related Acremonium strains. Curr Genet 19: 73–76
Weigel BJ, Burgett SG, Chen VJ, Skatrud PL, Frolik CA, Queener SW & Ingolia TD (1988) Cloning and expression in Escherichia coli of isopenicillin N synthetase genes from Streptomyces lipmanii and Aspergillus nidulans. J Bacteriol 170: 3817–3826
Whiteman PA, Abraham EP, Baldwin FL, Fleming MD, Schofield CJ, Sutherland JD & Willis AC (1990) Acyl-coenzyme A:6-aminopenicillanic acid acyltransferase from Penicillium chrysogenum and Aspergillus nidulans. FEBS Lett 262: 342–344
Yourno J, Kohno T & Roth JR (1970) Enzyme evolution: generation of a bifunctional enzyme by fusion of adjacent genes. Nature 228: 820–824
Zhu J & Schiestl RH (1996) Topoisomerase I involvement in illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol 16: 1805–1812
Zolan ME (1995) Chromosome-length polymorphism in fungi. Microbiol Rev 59: 686–698
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gutiérrez, S., Fierro, F., Casqueiro, J. et al. Gene organization and plasticity of the β-lactam genes in different filamentous fungi. Antonie Van Leeuwenhoek 75, 81–94 (1999). https://doi.org/10.1023/A:1001861025070
Issue Date:
DOI: https://doi.org/10.1023/A:1001861025070