Abstract
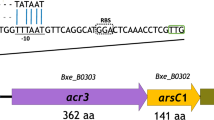
We reported earlier about the detection of a chromosomally located arsenic operon (arsRBC) in a gram-negative bacterium Pseudomonas fluorescens strain MSP3, which showed resistance to elevated levels of sodium arsenate and sodium arsenite. The genes for arsenic resistance were cloned into the HindIII site of pBluescript vector producing three clones MSA1, MSA2 and MSI3 conferring resistance to sodium arsenate and arsenite salts. They were further sub-cloned to delineate the insert size and the sub-clones were designated as MSA11, MSA12 and MSI13. The sub-clone pMSA12 (2.6 kb) fragment was further packaged into EcoRI-PstI site of M13mp19 and sequenced. Nucleotide sequencing revealed the presence of three open reading frames homologous to the arsR, arsB and arsC genes of arsenic resistance. Three cistrons of the ars operon encoded polypeptides ArsR, ArsB and ArsC with molecular weights ranging approximately 12, 37and 24 kDa, respectively. These polypeptides were visualized on SDS-PAGE stained with Coomassie blue and measured in a densitometer. The arsenic resistance operon (arsRBC) of strain MSP3 plasmid pMSA12 consists of 3 genes namely, arsR – encoding a repressor regulatory protein, arsB – the determinant of the membrane efflux protein that confers resistance by pumping arsenic from the cells and arsC – a small cytoplasmic polypeptide required for arsenate resistance only, not for arsenite resistance. ArsB protein is believed to use the cell membrane potential to drive the efflux of intracellular arsenite ions. ArsC encodes for the enzyme arsenate reductase which reduces intracellular As(V) (arsenate) to more toxic As(III) (arsenite) and is subsequently extruded from the cell. The arsenate reductase activity was present in the soluble cytoplasmic fraction in E. coli clones. In the context of specified function of the arsenic operon encoded proteins, uptake and efflux mechanisms were studied in the wild strain and the arsenate/arsenite clones. The cell free filtrates of the arsenate clones (MSA11 and MSA12) obtained from P. fluorescens containing the arsC gene showed that arsenate reduction requires glutathione reductase, glutathione (GSH), glutaredoxin and ArsC protein. The protein was purified in an active form and a spectrophotometric assay was developed in which the oxidation of NADPH was coupled to reduction of arsenate. The molecular weights and the location of the polypeptides were obtained from Coomassie stained SDS-PAGE of extracellular and intracellular fractions of the cells.
Similar content being viewed by others
References
Zhou T, Radaev S, Rosen BP & Gatti DL (2000) EMBO J. 19: 4838–4845
Dey S & Rosen BP (1995) Mechanisms in drug transport in prokaryotes and eukaryotes. In ‘Drug Transport in Antimicrobial and Anticancer Chemotherapy’ (Ed. by Georgopapadakou NH) Dekker, pp. 103–132, New York, NY
Rosen BP (1999) Trends Microbiol. 7: 207–212
Silver S & Ji G (1994) Environ. Health. Perspect. 102 (Suppl.3), 107–113
Cervantes C, Ji G, Ramirez J L & Silver S (1994) FEMS Microbiol. Rev. 15: 335–367
Silver, S & Keach D (1982) Proc. Natl. Acad. Sci. USA 79: 6114–6118
Xu C, Zhou L, Kuroda M & Rosen BP (1998) J. Biochem. 123: 16–23
Rosen BP, Dey S, Dou D, Ji G, Kaur P, Ksenzenko MY, Silver S & Wu J (1992) Ann. N.Y. Acad. Sci. 671: 257–272
Silver S, Ji G, Broer S, Dey S, Dou D & Rosen BP (1993) Mol. Microbiol. 8: 637–642
Ji G & Silver S (1992). J. Bacteriol. 174: 3684–3694
Prithivirajsingh S, Mishra SK & Mahadevan A (2001) Biochem. Biophys. Res. Comm. 280: 1393–1401
Silver S (1996) Gene 197: 9–19
Nakamuro K & Sayato Y (1981) Mutat. Res. 88: 73–80
Bergey's Manual of Determinative Bacteriology (1994) (Eds. by Holt JG, Krieg NR, Sneath PHA, Staley JL and Williams ST) Williams and Wilkins, Baltimore.
Hatch WR & Ott WL (1996) Anal. Chem. 40: 2085–2087
Bradford MM (1976) Anal; Biochem. 72: 248–256
Laemmli UK (1970) Nature 227: 229–230
Read RR & Consterton (1987) Can. J. Microbiol. 33: 1080–1090
Funahashi H, Maehara M, Yoshida T & Tagachi H (1987) J. Chem. Eng. Japan 20: 16–28
Ji G & Silver S (1992) Proc. Natl. Acad. Sci. USA 89: 9474–9478
Gladysheva TB, Oden KL & Rosen BP (1994) Biochemistry 33: 7287–7293
Ji G, Graber EA, Armes LG, Chen CM, Fuchs JA & Silver S (1994) Biohemistry 33: 7294–7299
Silver S & Phung Le T (1996) Annu. Rev. Microbiol. 50: 735–789
Sofia H J, Burland V, Daniels DL, Dluncett G & Blattner FR (1994) Nucleic Acids Res. 22: 2576–2586
Carlin A, Dey SW & Rosen BP (1995) J. Bacteriol. 177: 981–986
Diorio C, Cai J, Marmor J, Shinder R & DuBow MS (1995) J. Bacteriol. 177: 2050–2056
Takemaru KI, Mizuno M, Sato T, Takeuchi M & Kobayashi Y (1995) Microbiology 141: 323–327
Hedges RW & Baumberg S (1973) J. Bacteriol. 115: 459–460
Mobley HLT, Silver S, Porter FD & Rosen BP (1984) Antimicrobiol. Agents Chemother. 25: 157–161
Novick RP & Roth C (1986) J. Bacteriol. 95: 1335–1342
Summers AO & Jacoby GA (1978) Antimicrobid. Agents Chemother. 13: 637–640
Silver S, Budd K, Leathy KM, Shaw WV, Hammond D, Novick RP, Willsky GR, Malamy MH & Rosenberg H (1981) J. Bacteriol. 146: 983–996
Broer S, Ji G, Broer A & Silver S (1993) J. Bacteriol. 175: 3480–3485
Rosentein R, Niloleit K & Gotz F (1994) Mol. Gen. Genet. 242: 566–572
Mobley HLT & Rosen BP (1982) Proc. Natl. Acad. Sci. USA 79: 6119–6122
Chen CM, Misra TP, Silver S & Rosen BP (1986) J. Bacteriol. 261: 15030–15038
Wu JH & Rosen BP (1993) J. Biol. Chem. 268: 52–58
Wu JH & Rosen BP (1993) Mol. Microbiol. 8: 615–623
Dey S, Dou D, Tisa LS & Rosen BP (1994) Arch. Biochem. Biophys. 311: 418–424
Tisa LS & Rosen BP (1991) J. Biol. Chem. 265: 190–194
Dey S & Rosen BP (1995) J. Bacteriol. 177: 385–389
Dou D, Dey S & Rosen BP (1994) Antonie van Leeuwenhoek 65: 359–368
Rosentein R, Peschal A, Wieland B & Gotz F (1992) J. Bacteriol. 174: 3676–3683
Liu J & Rosen BP (1997) J. Biol. Chem. 272: 21084–21089
Westenberg DJ & Guerinot ML (1997) Adv. Genet. 36: 187–239
Sata T & Kobayashi Y (1998) J. Bacteriol. 180: 1655–1661
Butcher BG, Deane SM & Rawlings DE (2000) Appl. Env. Microbiol. 66: 1826–1833
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Prithivirajsingh, S., Mishra, S.K. & Mahadevan, A. Functional analysis of a chromosomal arsenic resistance operon in Pseudomonas fluorescens strain MSP3. Mol Biol Rep 28, 63–72 (2001). https://doi.org/10.1023/A:1017950207981
Issue Date:
DOI: https://doi.org/10.1023/A:1017950207981