Abstract
The thyroid gland is the crucial organ in the human body, secreting two hormones that help to regulate the human body’s metabolism. Thyroid disease is a severe medical complaint that could be developed by high Thyroid Stimulating Hormone (TSH) levels or an infection in the thyroid tissues. Hypothyroidism and hyperthyroidism are two critical conditions caused by insufficient thyroid hormone production and excessive thyroid hormone production, respectively. Machine learning models can be used to precisely process the data generated from different medical sectors and to build a model to predict several diseases. In this paper, we use different machine-learning algorithms to predict hypothyroidism and hyperthyroidism. Moreover, we identified the most significant features, which can be used to detect thyroid diseases more precisely. After completing the pre-processing and feature selection steps, we applied our modified and original data to several classification models to predict thyroidism. We found Random Forest (RF) is giving the maximum evaluation score in all sectors in our dataset, and Naive Bayes is performing very poorly. Moreover selecting the feature by using the feature importance method RF provides the best accuracy of 91.42%, precision of 92%, recall of 92% and F1-score of 92%. Further, by analyzing the characteristics and behavior of the dataset, we identified the most important features (TSH, T3, TT4, and FTI) of the dataset. In terms of accuracy and other performance evaluation criteria, this study could advocate the use of effective classifiers and features backed by machine learning algorithms to detect and diagnose thyroid disease. Finally, we did some explainability analysis of our best classifier to understand the internal black-box of our machine learning model and datasets. This study could further pave the way for the researcher as well as healthcare professionals to analyze thyroid disease in real time applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
As stated by the World Health Organization, thyroid illness is the most widespread endocrine disorder in the world after diabetes (https://www.who.int/) [1]. Hyperthyroidism and hypothyroidism are the most frequent thyroid gland illnesses, which have been recorded in more than 110 countries throughout the world, putting 1.6 billion people in danger, and a majority of these are found in Asia, Africa, and Latin America [2]. Currently, over 25,000 emergency clinics around the globe collect information on patients in various configurations. However, studies are conducted by traditional examination and measurable tests using the traditional method [3], which is time-consuming and costly. Doctors believe that, early disease detection, diagnosis, and treatment are critical in inhibiting disease development or even passing away. Despite numerous trials, clinical diagnosis is frequently regarded as a difficult task [4]. The thyroid is a tiny, butterfly-shaped gland that sits right below Adam’s apple at the base of the neck [5].The endocrine system is a complicated network of glands that controls the organization of many of the actions of the human body. The thyroid gland yields hormones that govern the human body’s metabolism. The most common cause is a lack of iodine; however it can also be caused by other circumstances [6]. T3, T4, and Calcitonin are the three hormones produced by the thyroid gland where T3 and T4 are just in the strictest sense [7]. Iodine is required for the creation of both hormones. We must receive this trace element through our diet because our systems cannot produce it. Iodine is absorbed into our bloodstream by food in our intestines and finally produces thyroid hormones. Hypothyroidism (underactive thyroid) is a malfunction in which the thyroid gland does not produce enough specific hormones [8]. A few symptoms had been seen early on in the course of hypothyroidism. Without giving much con- centration to hypothyroidism, this could be led to obesity. Moreover, several other problems like joint pain, heart disease, and sometimes infertility might be seen among patients [9]. Hyperthyroidism is a malfunction in which the thyroid gland produces many thyroid hormones that circulate in the blood- stream. Some symptoms of hyperthyroidism are nervousness, impatience, and increased hunger [10]. For thyroid prediction at early stage, we could use ma- chine learning, an area of computer science that has exploded in popularity in recent years and is likely to continue to do so in the future. A machine learning algorithm has several advantages, including high parallelism, speed, self-learning and noise error tolerance [11]. Machine learning allows humans to get insight from vast amounts of data that would otherwise be too difficult or impossible to process. By building a machine learning model, we can predict hypothyroidism and hyperthyroidism with the help of symptoms of the patient, which is a cost-effective and time-saving approach. The machine learning model is trained using data from various databases as input. It can be used to produce predictions for other input data once it has been trained. Several supervised machine learning algorithms are available in the literature [12,13,14,15,16]. We employed a Decision tree classifier, Random Forest Classifier, Gradient Boosting Classifier, Naive Bayes Classifier, K-Nearest Neighbor, Logistic Regression, and Support Machine Vector to predict thyroid disease in our study, and we could relate the performance of the algorithms to discover the finest method for more correctly predicting thyroid disease. In the end, a post hoc technique known as explainable artificial intelligence is used to understand and believe the output and outcomes produced by the black box machine learning algorithms.
The primary goal of this research is
-
To find a reliable machine learning classification method for predicting thyroid disease using the fewest possible features.
-
Identifying the most important feature to detect the thyroid disease
-
Validating the experimental results using explainable AI.
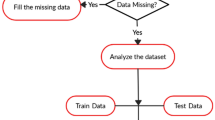
We also determined the most important aspects of our datasets for pre- dicting thyroid illness. Finally, we believe this research might significantly impact the scientific community for better understanding and applying ma- chine learning in the medical field especially thyroid disease prediction. The overall workflow of this study is depicted in Fig. 1. Firstly, we collect preprocessed, cleaned and resampled the data. Following that, two sub dataset was generated using the original dataset, which was used to predict thyroid Over- all, we followed seven steps for building the thyroid prediction model. Finally, validated our results using explainable AI.
2 Literature Review
Several works have been done so far in this relevant work. In [17], authors employed Decision Tree, Support Vector Machine, Artificial Neural Network, and the K-Nearest-Neighbor algorithm, among other classification algorithms, based on the dataset of thyroid gained from the UCI Repository (https://archive.ics.uci.edu/ml/ datasets/thyroid + disease), classification and prediction were performed, and accuracy was measured based on the output provided. Logistics Regression and SVM machine learning techniques to evaluate the Thyroid Dataset and RMS error, Precision, Recall, F1 measure, and ROC were used to compare these two methods in [18]. According to them, successful classifier was found to be logistic regression. Awasthi and Anil Antony [19] discussed using KNN, support vector machine (SVM), and machine learning algorithms to categorize and detect thyroid illness. They employed the K-nearest neighbor technique to approximate missing values in user input for thyroid diagnosis. A classification system for two categories of thyroid disease: hyperthyroidism and hyperthyroidism was proposed in [20]. During the preprocessing stage, missing values that are not a numerical constraint are identified, and the mean value of the matching column is used to fill in the gaps. The differential evolution technique is used to create child subsets from parent records. In [21], the authors used SVM as a classifier to distinguish thyroid disease. This investigation is based on two datasets. For classification, the authors employed Naive Bayes and support vector machines in [22]. Several grouping algorithms, like the K-nearest neighbor, support this idea. The Rapid miner device was used to conduct the research, and the findings reveal that K-nearest neighbor is more optimum than Naive Bayes in diagnosing thyroid issues. The K-nearest neighbor classifier was the most reliable, with accuracy of 93.44%, while the Naive Bayes classifier had only 22.56%. In [23], SVM surpassed K-Nearest Neighbor and Bayesian with an accuracy of 84.62 percent. KNN independently discovered the closest neighborhood. In [24], the authors have proposed several Thyroid prediction strategies based on data mining techniques. They investigated the link between T3, T4, and TSH, as well as hyperthyroidism and hypothyroidism. In addition to this, recently some other authors also use different machine learning modification technique to predict thyroid [25, 26]. Moreover, in [27] authors used different feature engineering method like forward backward and bidirectional feature elimination method for thyroid classification. In the area of healthcare, Pawar et al. [28] employed the XAI technique to model integrity, openness in feature selection, result monitoring, and model refinement. The same author also employed explainable AI in 2021 to offer a tool for comprehending machine learning models in the healthcare industry. To the best of our knowledge, Arjaria et al. in 2022 [29] used XAI to forecast the accuracy of decision tree algorithms with an explanation of key features, improving the models’ accuracy, and making the models more accountable by requiring them to explain the reasoning behind each decision. Despite the widespread application of AI and machine learning in the fields of medicine and diagnostics, we observed that there is still a gap in the ability of explainable machine learning to predict thyroid disease. In this study, we first predict the best ML model and then utilize XAI to analyze the best ML model’s “black box” for classifying thyroid diseases.
3 Material and Methods
3.1 Dataset Description
The first natural step towards the development of a machine learning model is the collection of data. The data was taken from the UCI (https://archive.ics.uci.edu/ml/datasets/Thyroid+Disease) machine learning repository [30]. We took three datasets (hypothyroid, hyperthyroid, and sick) from the UCI machine learning repository and combine them to create our final dataset, which has 3221 entries.
There is a total of 30 features; six of the features are real number properties, while the remainder is category traits, as shown in Table 1. Pre-processing is done to improve the quality of the dataset obtained for further analysis. The histogram of all the attributes is visualized in Fig. 2 after dropping the two attributes (TBG measured and TBG) because of the sizeable missing value.
Visual representation of the dataset for all patients with all 28 attributes. The frequency distribution of the dataset can now be easily seen from the histogram. Overall the attributes age, TSH, T3, T4, T4U, and FTI appeared to be normally distributed. Whereas, other qualities are categorical in nature
3.2 Data Pre-processing
Raw data from the real world is frequently incomplete, unreliable, and devoid of specific behaviors or trends. They are also likely to have many mistakes in them [31]. As a result, they are pre-processed into a format that the machine learning algorithm can use for the model once they have been collected. The data pre-processing phase should be given much attention in order to get the best model quality. It includes several tasks employed in the process to make the data more relevant. In this study, we followed the following steps in order to preprocess the data.
At first, many unclear values did not have any significant meaning. So, we removed those unclear values to get better results from this process by reducing the attributes of the dataset. Following that, we replaced the missing value because our dataset had so many missing values. We took different steps to handle missing values, for example, filling missing values with median and mode. In addition, the categorical data was encoded into integer format so that data with transformed category values may be fed into models to improve prediction accuracy. Furthermore, we handled the imbalanced data from our datasets in which the target class had an unequal distribution of observations. For balancing our dataset, we employed the resampling technique. Finally, we spilled the datasets into training and test sets. The training dataset was utilized to fit the model, and test sets were used to make predictions and compare them to the predicted values. In this study, 70 out of 100 data was used for training, and 30 out of 100 was used for testing.
3.3 Feature Selection Methods
Feature selection is a strategy for limiting the input variable to the model by removing insignificant data and only using valuable data [32]. The purpose of feature selection in machine learning is to determine the best set of characteristics for building effective models of the phenomena being studied. In this study, for selecting the most important feature, we used the univariate feature selection approach and the feature importance method [33].
3.4 Selection of the Classification Algorithms
Before selecting an algorithm, there are a few things to remember, the size of the training data, the output’s accuracy and/or interpretability, time spent on training or speed, linearity, and the number of features [34]. In this investigation, we took seven popular machine learning classification algorithms for solving this dataset because we are trying to figure out which algorithm performs better on our dataset. In order to predict the thyroid, we use Decision Tree Classifier, Random Forest Classifier, Gradient Boosting Classifier, Naive Bayes Classifier, Logistic Regression, K-Nearest Neighbor, and Support Vector Machine (SVM) algorithms.
Supervised Machine Learning algorithms like Decision Trees are typically used to tackle classification and regression issues by splitting the data based on specific criteria. While the data is divided among the nodes, the final decision is provided by the leaves. The problems with this method are over-fitting, although Random Forest offers a solution that is based on an ensemble modeling strategy. The naive Bayes algorithm is based on conditional probability and use a probability table as the model. Throughout the training session, the table is updated. Due to its effectiveness, performance, adaptability for modest amounts of training data, dealing of both discrete and continuous data, and capability to address binary and multi- class classification challenges, the technique has benefits over competing methods. However, because NB models are too simplistic, properly trained and optimized models frequently outperform them.
Computational efficiency, ease of regularization, and simplicity in implementation are some benefits of Logistic Regression. However, its inability to tackle non-linear problems, susceptibility to overfitting, and poor performance until all independent variables are recognized may sometimes causes problem of using this algorithm. Unsupervised K Means Clustering is a widespread choice for clustering problems. When variables are large, it is computationally more effective than hierarchical clustering. The algorithm’s order of complexity, making it computationally efficient. However, K value prediction is challenging and the performance of globular clusters is compromised.
We can use SVM for both classification and regression problem. The decision boundary, a hyperplane is required to divide a collection of objects into their many classes. It can manage structured and semi-structured data. Moreover, it can manage complex functions if the right kernel function can be determined. SVM has a lower likelihood of over fitting. With a huge data collection, though, its performance suffers because of the longer training times.
3.5 Evaluation of the Model
In machine learning, performance metrics refer to how well an algorithm per- forms depending on various criteria such as precision, accuracy, recall, and F1 score [35,36,37]. The following sections go through several performance metrics.
3.5.1 Accuracy
The percentage of correct test data predictions referred to as accuracy. It is easy to calculate by dividing the number of forecasts by the number of correct guesses. The formula for calculating the accuracy is given below:
3.5.2 Precision
The precision score is used to assess the model’s correctly counting genuine positives among all positive predictions. The following is the formula for calculating precision:
3.5.3 Recall (Sensitivity)
The recall score used to assess the model’s performance in terms of accurately counting true positives among all actual positive values. Below is the formula for determining the recall.
3.5.4 F1 Score
The F1-score is the harmonic mean of precision and recall score, and utilized as a metric in situations when choosing either precision or recall score can result in a model with excessive false positives or false negatives. The F1 score measured as follows.
After combining three datasets, our final thyroid dataset had 3221 number of instances of 3221 patients. Along with the target value, we had 30 attributes. There were no missing values in our data. When we looked back at the original dataset, there were missing values in several columns.’nan’ is used to replace these values. Then we convert it into the numerical format. Because the missing values, except sex, are from numeric attributes, they are replaced with the median value of the respective columns. However, sex is a categorical attribute, and the missing value of it is replaced with a mode value of the respective attribute. Initially, we dropped two attributes, TBG and TBG measured, as the majority of values of these attributes are missing. Because the majority of data of these columns are missing. Our categorical attribute is mapped to numeric values, done manually with programming. For converting those values into numeric values, we use a label encoder. Our other attributes are in the form of objects. As a result, we convert them to integer format to fit them into our model. Our dataset is imbalanced because the target class has an uneven distribution of observations. There are 2753 observations under the negative class label, 220 observations under the hypothyroid class label, 171 observations under the sick class label, and 77 observations under the hyper- thyroid class label. When dealing with unbalanced datasets, typical machine learning methods may create biased, erroneous, and unsatisfactory classifiers.
Standard classifier methods favor classes with many instances, such as Decision Tree and Logistic Regression. Typically, they can only anticipate data from the vast majority of classes. The minority class’s traits are frequently dismissed as noise and ignored. As a result, the minority class has a higher chance of being misclassified than the majority class. Because Machine Learning Algorithms are typically design to improve accuracy by reducing error, this occurs. Therefore, we convert the dataset into a balanced one to obtain the desired result. We use a resampling technique to ensure that the minority and majority classes are equal. Finally, the distribution of observations in our dataset is even across our entire class.
3.6 Explainable Artificial Intelligence
Explainable artificial intelligence (XAI) is a collection of techniques and strategies that eventually enable human users to grasp and trust the output and results generated by the black box machine learning methods. In this study, a post hoc XAI approach has been considered to explain the model. The post hoc approaches analyze the model after training, but they do so without restricting the model's complexity. Therefore, the explainability does not affect the performance of the model. The complexity of the machine learning model is, however, restricted through the use of intrinsic approaches. Again, based on the scope, there can be two types of explainability: global and local. A global explanation of a machine learning (ML) model specifies which features are vital to the overall model's outcome. In contrast, a local approach only explains single data points. In this study, we used Shapley additive explanations (SHAP) [38]and local interpretable model-agnostic explanations (LIME) [39] for the global and local explanations, respectively.
For N number of explanatory variables, in terms of local accuracy each prediction made by the SHAP method is approximated by f(x) with g(x′), and a quantity ϕj ∈ R. Which can be defined as follows[40]:
Three major properties of SHAP Local accuracy, missingness, and consistency can only be satisfied by one explanatory model defined by as follows:
where z′ ∈ {0,1} N: binary variable’s linear function, z′\j representing setting at zi′ = 0, and non-zero entities is denoted by |z′|.
Whereas LIME attempt to fit a local model with sample data points that resemble the observation being addressed. Thus, each observation x of LIME can be obtained by as follows [40]:
where locality aware loss L, potentially interpretable models is denoted by Q, πx(z): distance between an instance z and x, and ψ(q): A metric for the explanation's complexity q ∈ Q.
4 Result and Discussion
A comparison of seven different machine-learning algorithms was conducted in this study. Decision Tree Classifier, Random Forest Classifier, Naive Bayes Classifier, Gradient Boosting Classifier, Logistic Regression Classifier, K- Nearest Neighbor, and Support Vector Machine were utilized for thyroid disease prediction. Firstly, we collect and preprocessed the data and then fed the data to train the model. By comparing the scores, various performance criteria, including accuracy, precision, recall, and F1-score, are utilized to establish whether an algorithm is superior to others. We divide our dataset into three formats: the first set considering all attributes, the second set with 14 feature selection process attributes, and the third with 14 univariant feature selection process attributes. We narrowed down attributes based on their correlation with the target, which we calculated with the feature selection process and univariant feature selection methods. Overall, the results of various algorithms are explained in the next part of this result analysis.
4.1 Descriptive Statistics of the Dataset
Exploratory data analysis (EDA) is a sort of data analysis that employs data visualization to evaluate and investigate data sets and describe their key properties [41, 42]. EDA is mainly used to examine what data might reveal outside formal modeling or hypothesis testing tasks and to understand variables and their interactions better. It can also help us to figure out if the statistical methods we are contemplating for data analysis are appropriate. Our dataset has 28 attributes, with only six of them being numeric. Therefore, we give a short descriptive statistic of our dataset in Table 2. We can see that all of the attributes have 3221 values in this table. So, before we train the model, we use various techniques to fill in the missing values. We can also see that the average age of the patients is 52.4, implying that the most patients were elderly.
The youngest person was one year old, and the oldest person was 94 years old. The age distribution of the data is skewed, indicating that the population with a low age is absent. The standard deviation is 19.1, indicating the sparse- ness of the age group, which ranges from 57 to 73 years old. TSH mean was 6.322 mIU/L, indicating that most patients’ TSH levels were not expected. TSH levels should be between 0.5 and 5.0 mIU/L to be considered normal. TSH had a minimum value of 0.005 mIU/L and a maximum value of 478.0 mIU/L. The mean T3 value was 1.95 nmol/L, with a minimum of 0.05 nmol/L and a maximum of 10.6 nmol/L. The mean value of TT4 is 107.55. The maximum value of TT4 is 430 and the minimum of TT4 is 2.
In the case of T4U, the mean value is 0.988 mIU/mL. The maximum value of T4U is 2.12 mIU/mL and the minimum value of T4U is 0.31 mIU/mL. More- over, the mean value of FTI is 110.26. The correlation between all the numeric data is depicted in Fig. 3. Figure 3 shows that TT4 and FTI have a strong relationship. We can get a better understanding of this correlation table if we look at the heat map. Fig. 4 depicts a heatmap of all attribute correlations. Form the heatmap and numerical correlation of the above figure, we can draw interpretation about the correlation among the variable. It is clear from the heatmap that T4U measured and FTI measured has very strong correlation. Moreover some other parameter’s also visualized very strong relationship like TT4 with T3, T4U with FTI, TT4 with FTI and TTI with T4U.
4.2 Category Class Blanching
The target class has an uneven distribution of observations, which makes our dataset unbalanced. There are 2753 observations under the negative class label, 220 observations under the hypothyroid class label, 171 observations under the sick class label, and 77 observations under the hyperthyroid class label. So, our dataset is highly unbalanced.
As a result, machine learning classifiers faced some difficulties in making accurate predictions on our dataset. Because classic classifiers such as Decision Tree and Logistic Regression favor classes with many occurrences, they typically only forecast data from the vast majority of classes. The features of the minority class are frequently rejected and treated as noise. The graphical representation of our classes is shown in Fig. 5.
We can see that our dataset is entirely skewed. We focus on balancing the classes in the training data before delivering the data as input to the classification model. The primary purpose of class balancing is to either increase the frequency of the minority class or lower the frequency of the majority class. This is done to ensure that the number of instances in both classes is about equal. We employed the resampling technique to balance our dataset. Resampling is a common strategy for dealing with very imbalanced datasets. Under-sampling involves deleting samples from the majority class and/or introducing additional examples from the minority class. All our classes have an equal number of 2753 observations. The balanced plot is shown in Fig. 6.
After resampling, we found our final balanced dataset. Now we can build our model using this dataset which will give us a more accurate result. After resampling, we have a total of 11012 instances
4.3 Performance Analysis of Different Algorithm
Our original dataset, which included all features, was first utilized to evaluate several machine learning measures. After that, we used our balanced dataset to test multiple machine-learning models. This study selected the dataset’s important features using feature importance methods and univariate feature selection techniques. In our experiments, those vital features are then used to identify the model’s precision, accuracy, recall, and F1 score.
The data we use is typically divided into two categories: training data and test data. In this study, 70% of the data was utilized for training and 30% for testing. So, out of our 11,012 dataset instances, 7708 were used for the training set. 3304 of the 11,012 dataset instances were used in the testing set. Using the testing, we can determine the accuracy of our model and how well it can predict thyroid disease. We used the Sklearn library to split our data set as a train and test set. Sklearn, model selection train, and test split library component, split the dataset randomly with specified portion, and we get the random train and test part from the entire dataset. After training the model with all algorithms, the testing dataset was used to test the methods. The F1-score, recall, precision, and accuracy were used to evaluate the model’s performance. The entire study aimed to see which algorithm could best classify diseases.
This section highlights the study’s outcomes and introduces the top performer based on several performance criteria. At first, performance was measured using our raw dataset. Secondly, performance was measured using a dataset containing 14 attributes derived from the feature importance method. Third, performance was determined by considering 14 attributes from the univariate feature selection. Finally, we compare various performance metrics of various algorithms and feature categories.
4.3.1 Results Using All Features
We apply the selected algorithms to our dataset. Our dataset has 28 attributes; among them, the category is the target. The algorithms are then compared using various performance metrics. We can see from Table 3 that the Logistic Regression algorithm has the highest accuracy of any algorithm. After Logistic Regression, Support Vector Machine, Gradient Boosting Classifier, and Decision Tree Classifier have higher accuracy. Predictor accuracy refers to how well a predictor can forecast the value of a predicted characteristic for fresh data. In contrast, classifier accuracy refers to a classifier’s ability to predict the class label correctly. However, accuracy does not always provide good performance metrics to compare algorithms, so consider other metrics, for instance, recall, precision, and F1 score. We now assess our model’s performance using various metrics such as recall, precision, and F1 score.
Logistic Regression, as shown in Table 3, outperforms in terms of accuracy. However, this algorithm’s precision, recall, and F1 score are all low. We got an accuracy of 84.48%, precision of 25%, recall of 24%, and F1 score of 25 from Logistic Regression, which outperforms the other six classification algorithms for this dataset. The Support Vector Machine, Gradient Boosting Classifier, and Decision Tree Classifier perform as well. However, precision, recall, and F1-score are all extremely low in each case. As a result, we can only measure them using accuracy. However, accuracy cannot always provide us with an accurate measure of performance. Random Forest has a 74.4% accuracy, but precision, recall, and F1 score are all low. The accuracy of the K-Nearest Neighbor is 72.18 percent. On the other hand, Naive Bayes gives us a low score for this experiment. This algorithm only has a 16.44 percent accuracy, which is highly unsatisfactory. From the result, we can also say that Logistic Regression gives us the best prediction for our dataset. Naive Bayes gives us the poorest prediction in this case. As a result, we can conclude that for our dataset, Logistic Regression is the best classification algorithm, while Naive Bayes is the worst.
4.3.2 Results for Our Dataset Using Feature Importance Method
We determine our 14 best-correlated features from our dataset using the feature importance technique. We apply the seven algorithms to the 14 features chosen using the method. The algorithms are then compared using various performance metrics. All the selected features are presented in Fig. 7 with their importance value.
We apply Random Forest Classifier, Decision Tree Classifier, Gradient Boosting Classifier, Naive Bayes Classifier, Logistic Regression Classifier, K-Nearest Neighbor, and Support Vector. We can see from the above bar chart that the Random Forest algorithm outperforms all others in terms of accuracy. After Random Forest, Decision Tree Classifier and Gradient Boosting Classifier have higher accuracy. As previously stated, accuracy is not always an appropriate metric when comparing algorithms, so consider alternative metrics like precision, recall, and F1-score. The performance metrics of all seven algorithms are listed in Table 4.
Random Forest beats all other performance criteria, such as accuracy, pre- cision, recall, and F1 score, as seen in the table above. We have the highest accuracy of 91.92 percent, the highest precision of 92 percent, the highest recall of 92 percent, and the highest F1 score of 92 percent. So, for our dataset with 14 feature importance attributes, Random Forest outperforms the other six classification algorithms. Following that, the Gradient Boosting and Decision Tree Classifier perform admirably. However, both the Decision Tree Classifier and the Gradient Boosting Classifier have the same precision, recall, and F1 score. Moreover, in the case of Gradient Boosting, accuracy is improved. So, in terms of accuracy, we can say that Gradient Boosting outperforms Decision Tree Classifier. K-Nearest Neighbor has an accuracy of 86.22 percent and an F1 score of 86 percent. With a 73.7 percent F1 Score, SVM provides 73.7 per- cent accuracy. With an F1 score of 86 percent, Logistic Regression has a 73.15 percent accuracy. Finally, Naive Bayes gives a 64 percent F1 score and 67.86 percent accuracy, respectively. The confusion matrix tells us how accurate the classifier is at making predictions. The confusion matrix of all seven classification algorithms is shown in Fig. 8.
From the confusion matrix, as shown in Fig. 8, we can also say that Random Forest gives us the best prediction, and Naive Bayes gives us the poorest prediction in this case. As a result, we can conclude that for our chosen dataset, Random Forest is the best classification algorithm.
4.3.3 Results for Our Dataset Using Univariate Feature Selection Method
In this case, we use the univariate feature selection method to select our important features. The top 14 features with their correlated score with our target are given in Fig. 9. We apply the Decision Tree Classifier, Random Forest Classifier, Gradient Boosting Classifier, Naive Bayes Classifier, Logistic Regression Classifier, K-Nearest Neighbor, and Support Vector to the selected features. We can observe from Table 5 that our results are slightly different from previous results. Random Forest provides the best accuracy of 90.4 percent this time as well. After Random Forest, Decision Tree Classifier and Gradient Boosting Classifier have higher accuracy. Decision Tree Classifier and Gradient Boosting Classifier both have an accuracy of 89.55 percent and 89.35 percent, respectively. K Neighbors has an accuracy rate of 86.07 percent. The accuracy of SVM in- creased to 74.5 percent, whereas that of Logistic Regression decreased to 71.82 percent. Besides, the accuracy of Naive Bayes fluctuates a lot for this dataset. As a result, we conclude that this method is ineffective compared to the feature importance technique. Other performance metrics of all seven algorithms on this dataset are also presented in Table 5.
Table 5 shows that the performance metrics differ significantly from the previous test result. Logistic Regression, K Neighbors, and Support Vector Machine all have the same precision. The precision of the Decision Tree Classifier, Random Forest Classifier, Gradient Boosting Classifier, and Naive Bayes Classifier, on the other hand, decreases. K-Neighbors, SVM, and Logistic Regression all have the same recall. On the other hand, the recall of the Decision Tree Classifier, Random Forest Classifier, Gradient Boosting Classifier, and Naive Bayes Classifier falls. F1-Score provides a comprehensive view of precision and recall simultaneously, as shown by the fact that the F1-Score is the same for Logistic Regression, K Neighbors, and SVM. The F1 Score of Naive Bayes decreases. So, based on the table above, we can conclude that Random Forest is the best performer. After that, the Decision Tree Classifier performs admirably. Gradient Boosting Classifier and Decision Tree Classifier are nearly equal in this race, but Decision Tree Classifier outperforms Gradient Boosting Classifier by a small margin. However, Naive Bayes reduces performance across the board. The confusion matrix of all the seven classification algorithms is shown in Fig. 10.
We can also conclude from the confusion matrix that Random Forest pro- vides the best prognosis. In this case, Nave Bayes gives us the worst prediction. Overall results with all classifiers and features in this investigation are depicted in Fig. 11.
4.4 Explainability Analysis
We have achieved the best results for the Random Forest model, so we only explore the explanation for that model. Shapley additive explanations (SHAP) is a game theory-based method for interpreting the findings of machine learning models. It provides a procedure for determining and displaying the comparative importance of individual features of the model. This method approximates the individual contribution of each feature for each instance in the dataset. The importance of the feature is then assessed by analyzing the model’s results with and without the feature. To explain the model, we have generated four different visualizations of the feature effectiveness for the four classes. From Fig. 12, the contribution of the feature for the hypothyroid class can be interpreted. The majority of the instances have been classified as hypothyroid, where the TSH test value and T3 test value have a comparatively higher value. On the other hand, the lower values of the TT4 test and referral source significantly impact the predicted hypothyroid class for some instances. Figure 13 provides the important features for the explanation of the sick class. The figure also shows that the T3 feature has the highest feature importance for the Sick class. However, if the value of the T3, THS, and TT4 tests are low, then the model predicts a patient as sick. For a small number of instances, the higher values of the FTI feature is crucial for being classified as sick. Figure 14 illustrates that the FTI and T4U measured are the most important features for a patient to be classified as negative. The instances that have the normal range of the FTI test value are classified as negative. Figure 15 shows that the FTI and T3 test values are essential for the hyperthyroid class. It is interesting to observe that pregnancy can be a reason for hyperthyroid.
Local interpretable model-agnostic explanations (LIME) is an explainable AI technique that helps show how a machine learning model works and makes each forecast of the model easy to understand by itself. Since the method describes the classifier for a single instance, it works well for local explanations. LIME’s workings are based on the idea that any complicated model is simple on a regional scale. So, LIME tries to make a simplified model based on a single instance in the hopes that this model will mimic the behavior of the global model at that particular instance. The simplified model can then be used to figure out how that complex model is working.
We have used lime for the local explanation of our model. Using LIME, we can interpret a patient’s classification result, and Fig. 16 is one of them. The actual class of that patient was sick. The predicted class of the patient is also sick by our model. From the figure it can be explained why the instance is classified as sick. Figure 16a represents our model’s predicted probability for each class. Figure 16b represents the constraints fulfilled by that instance to be classified as sick. The model is interpreted as sick as the instance has test value T3 ≤ 1.20, FTI ≤ 103.00, TT4 ≤ 85, etc. Other important features, such as T4U, age, TSH, etc., pushed the instance to be classified as sick. Figure 16c shows the actual feature values of that instance.predicted class: hypothyroid
Figure 17 is another local explanation of an instance correctly classified as hypothyroid. From Fig. 17a, we can see the prediction probabilities. Figure 17b represents the conditions for which the instance has been classified as hypothyroid.
Our model classified both instances depicted in Figs. 18 and 19 as negative. Figure 19 illustrates the LIME explanation for an incorrectly categorized instance. The actual class of the instance is hyperthyroid, although the model predicts it as negative. Figure 19a represents the likelihood of prediction for each class. With a class probability value of 0.29, the actual class can be shown to be in second place. Figure 19b shows that several factors, like FTI, TT4, T3, etc., attempted to push the model’s output to the correct class. Nevertheless, Fig. 19c illustrates that the other factors such as lithium, TSH, T3, TT4, and so on pushed the model to the negative class. Notable is the fact that several criteria, such as TT4 > 122.00 and T3 > 2.20, are the same in the negative and hyperthyroid classes. However, for this instance, the other factors play an important role in predicting.
4.5 Discussion
As machine learning is using almost all aspect of data analysis. So practical implementation of machine learning model for medical data analysis especially for thyroid disease detection may save huge amount of expert physician requirements in this field. However medical data is very sensitive therefore perfect model is the basic requirements for medical data analysis. Selecting some appropriate feature data points as well as effective machine learning algorithm may pave the way for healthcare to automatic thyroid detection. In this study, we did feature engineering method to identify the best machine learning classification algorithm depending on the data feature used for thyroid detection. Furthermore, we validated our identified best performing model as well as the features which influenced most for the classification by XAI. It is clear from the performance of different algorithms that each algorithm performed better depending on whether a subset of features or full features were used. Depending on the situation, each algorithm has the inherent ability to outperform others. RF, for example, outperforms all other algorithms in terms of accuracy of 91.42% and 90.4% respectively in our dataset for the case of FS1 and FS2. Furthermore, we know that SVM performs better for small data sets, and ensemble-type classifiers like Random Forest perform better for large data sets. Missing values play a significant role in decision trees. Even after imputing, it cannot produce the same results as a perfect dataset. However, for our case, DT also performed very well of accuracy 90.43% in FS1. Another good classifiers was Gaussian Naive Bayes. However, it did not perform well with our dataset. The presumption that all attributes were independent was the reason for this. Results and Analysis would have been less accurate if there was a dependency between the features in the dataset. The accuracy of the K-Nearest Neighbor increases as the number of K we choose increases. It ensures that the given point and the dataset are similar. The performance of algorithms that use all the dataset’s features FS was poor relative to FS1 and FS2 for most of the algorithms. After reducing the attribute in the dataset, most of the cases machine learning algorithm performance improved. When there are many attributes, classifier algorithms become complicated, and prediction results vary. Because this is the standard process of evaluating algorithms, performance metrics after converting categorical values, balancing our dataset, and feature selection are used for dataset comparison. Therefore, by considering all of the situations and the performance of metrics used in this experiments we suggests that the Random Forest algorithm and FS1 features should be used to train the model to predict hypothyroidism and Hyperthyroidism more correctly. Furthermore, from the XAI analysis we could also observed that the feature attribute TSH, T3, TT4, FTI and T3 contributed the most to classify hypothyroid and hyper- thyroid can be obtained by using the feature importance method FS1. As we use the limited data points in our study there is still a chance for the biasness of the result specially for the case of using all feature to classify the thyroid. Moreover, we balance the dataset before final classification which might not always produced perfect data points. Exact clinical data points might further validate our approach which we are looking for.
With this UCI adjusted dataset, several studies [3, 28] have been conducted to determine the best suited machine learning model for thyroid categorization. Some other works [18, 21, 24] also noted for the prediction of thyroid disease using different dataset. As per our concern, in this paper, we apply XAI to verify the critical features from this dataset that led the best fitted model to predict specific classes using explainable artificial intelligence and our result is relatively comparable to the existing work.
5 Conclusion
After reducing the features using the feature importance technique and univariate feature selection technique, we tested our collected dataset on various machine learning classifiers to see which classifier gave us the best accuracy. After analyzing the data, we discovered that Logistic Regression outperforms all other classification algorithms for our dataset. When all features are considered, Logistic Regression yields an accuracy score of 84.48 percent. When we use the feature importance method to narrow down the feature set, the Random Forest Classifier gives an accuracy score of 91.92 percent. The accuracy of the Decision Tree Classifier and the Gradient Boosting Classifier is 90.5 percent and 90.43 percent, respectively. When we use the univariate feature selection technique to narrow down the feature set, Random Forest also gives the highest accuracy score of 90.4 percent. The second-best algorithm is the Decision Tree Classifier, which has an accuracy score of 89.55 percent; the third-best algorithm is the Gradient Boosting Classifier, which has an accuracy score of 89.35 percent. From explainability analysis, we can conclude that most instances have been classified as hypothyroid on the basis of the features TSH, T3 TT4. We can also identified that the FTI and T3 test values are important for the hyperthyroid class. So, the feature importance technique is more accurate than the univariate feature selection technique in determining correlated features. Thus, after looking at all of the performance metrics, we decided that the Random Forest Classifier, Decision Tree Classifier, and Gradient Boosting Classifier and feature importance technique might be a potential choice for predicting hypothyroidism and hyperthyroidism. Though we got relatively better result by feature engineering however there is still room for the search for more perfect model as well as dataset feature selection scheme to further improvement of the result. Moreover, we try our best to clear about biological term in this study however for any kind of our representational limitations we will improve in future.
Data Availability
This research study is based on an open-source dataset. The dataset can be accessed from this link: https://archive.ics.uci.edu/ml/datasets/Thyroid+Disease Thyroid Disease.
Abbreviations
- TSH:
-
Thyroid stimulating hormone
- XAI:
-
Explainable artificial intelligence
- UCI:
-
University of California, Irvine
- SVM:
-
Support vector machine
- KNN:
-
K-nearest neighbor
- T3:
-
Triiodothyronine
- T4:
-
Thyroxine
- ML:
-
Machine learning
- TT4:
-
Thyroxine hormone
- T4U:
-
Thyroxine utilization rate
- FTI:
-
Free thyroxine index
- TBG:
-
Thyroxine binding globulin
- NB:
-
Naive Bayes
- DT:
-
Decision tree
- TP:
-
True positive
- TN:
-
True negative
- FP:
-
False positive
- FN:
-
False negative
- LIME:
-
Local interpretable model-agnostic explanations
- SHAP:
-
Shapley additive explanations
- EDA:
-
Exploratory data analysis
- LR:
-
Logistic regression
- GB:
-
Gradient boosting
- FS1:
-
Feature Set 1
- FS2:
-
Feature Set 2
- FS:
-
Feature Set
References
Biondi B, Kahaly GJ, Robertson RP. Thyroid dysfunction and diabetes mellitus: two closely associated disorders. Endocr Rev. 2019;40(3):789–824.
Alam Khan V, Khan MA, Akhtar S. Thyroid disorders, etiology and prevalence. J Med Sci. 2002;2(2):89–94.
Sonu CE, et al. Thyroid disease classification using machine learning algorithms. J Phys. 2021;1963:12140.
Yasir Iqbal Mirut SM. Thyroid disease prediction using two tier ensemble classifier. Int J Adv Sci Technol. 2020;29:4460–71.
Bhaladhare V, Chouragade NB, Balpande D, Bhande A, Ambad RS, Bankar N. Ayurvedic management of hypothyroidism. Nat Volat Essen Oil J. 2021;1440–7.
Knudsen N, Laurberg P, Perrild H, Bulow I, Ovesen L, Jørgensen T. Risk factors for goiter and thyroid nodules. Thyroid. 2002;12(10):879–88.
Garg MK, Mahalle N, Kumar K. Laboratory evaluation of thyroid functions: dilemmas and pitfalls. Princ Pract Thyroid Gland Disord. 2017. https://doi.org/10.5005/jp/books/13094_4.
Feller M, Snel M, Moutzouri E, Bauer DC, de Montmollin M, Aujesky D, Ford I, Gussekloo J, Kearney PM, Mooijaart S, et al. Association of thyroid hormone therapy with quality of life and thyroid-related symptoms in patients with subclinical hypothyroidism: a systematic review and meta-analysis. JAMA. 2018;320(13):1349–59.
Unuane D, Velkeniers B. Impact of thyroid disease on fertility and assisted conception. Best Pract Res Clin Endocrinol Metab. 2020;34(4): 101378.
Abbas S. To determine the frequency of undiagnosed hyperthyroidism in patients presenting with generalized anxiety disorder. J Evol Med Dent Sci. 2013;2(8):930–8.
Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015;349(6245):255–60.
Choudhary R, Gianey HK 2017 Comprehensive review on supervised machine learning algorithms. In: 2017 International conference on machine learning and data science (MLDS), pp. 37–43. IEEE
Crisci C, Ghattas B, Perera G. A review of supervised machine learning algorithms and their applications to ecological data. Ecol Model. 2012;240:113–22.
Osisanwo F, Akinsola J, Awodele O, Hinmikaiye J, Olakanmi O, Akinjobi J. Supervised machine learning algorithms: classification and comparison. Int J Comput Trends Technol (IJCTT). 2017;48(3):128–38.
Praveena M, Jaiganesh V. A literature review on supervised machine learning algorithms and boosting process. Int J Comput Appl. 2017;169(8):32–5.
Singh A, Thakur N, Sharma A 2016 A review of supervised machine learning algorithms. In: 2016 3rd international conference on computing for sustainable global development (INDIACom), pp. 1310–1315. IEEE
Tyagi A, Mehra R, Saxena A 2018 Interactive thyroid disease prediction system using machine learning technique. In: 2018 fifth international conference on parallel, distributed and grid computing (PDGC), pp. 689–693. IEEE
Godara S, Kumar S. Prediction of thyroid disease using machine learning techniques. Int J Electron Eng. 2018;10(2):787–93.
Aswathi A, Antony A 2018 An intelligent system for thyroid disease classification and diagnosis. In: 2018 second international conference on inventive communication and computational technologies (ICICCT), pp. 1261–1264. IEEE
Geetha K, Baboo SS. An empirical model for thyroid disease classification using evolutionary multivariate bayseian prediction method. Global J Comput Sci Technol. 2016;16:1–9.
Kousarrizi, MRN, Seiti F, Teshnehlab M. An experimental comparative study on thyroid disease diagnosis based on feature subset selection and classification. IJECS. 2012;12:13–9.
Chandel K, Kunwar V, Sabitha S, Choudhury T, Mukherjee S. A comparative study on thyroid disease detection using k-nearest neighbor and naive bayes classification techniques. CSI Trans ICT. 2016;4(2):313–9.
Singh N, Jindal A. A segmentation method and comparison of classification methods for thyroid ultrasound images. Int J Comput Appli. 2012;50(11):43–9.
Begum A, Parkavi A (2019) Prediction of thyroid disease using data mining techniques. In: 2019 5th international conference on advanced computing & communication systems (ICACCS), pp. 342–345). IEEE
Almahshi HM, Almasri EA, Alquran H, Mustafa WA, Alkhayyat A 2022: Hypothyroidism prediction and detection using machine learning. In: 2022 5th international conference on engineering technology and its applications (IICETA), pp. 159–163 (2022). IEEE
Chaganti R, Rustam F, De La Torre Dıez I, Mazon JLV, Rodrıguez CL, Ashraf I. Thyroid disease prediction using selective features and machine learning techniques. Cancers. 2022;14(16):3914.
Alyas T, Hamid M, Alissa K, Faiz T, Tabassum N, Ahmad A. Empirical method for thyroid disease classification using a machine learning approach. BioMed Res Int. 2022. https://doi.org/10.1155/2022/9809932.
Pawar U, O’Shea D, Rea S, O’Reilly R 2020 Incorporating explainable artificial intelligence (xai) to aid the understanding of machine learning in the healthcare domain. In: AICS, pp. 169–180
Arjaria SK, Rathore AS, Chaubey G. Developing an explainable machine learning-based thyroid disease prediction model. Int J Bus Anal (IJBAN). 2022;9(3):1–18.
Dua, D., Graff, C.: Uci machine learning repository [https://archive.ics.uci.edu/ml]. irvine, ca: University of california, school of information and computer science. IEEE transactions on pattern analysis and machine intelligence (2019)
Kumar A, Tyagi AK, Tyagi SK. Data mining: various issues and challenges for future a short discussion on data mining issues for future work. Int J Emerg Technol Adv Eng. 2014;4(1):1.
Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–82.
Jovi´CA, Brki´CK, Bogunovi´CN 2015 A review of feature selection methods with applications. In: 2015 38th international convention on information and communication technology, electronics and microelectronics (MIPRO), pp. 1200–1205. Ieee
Cui S, Tseng H-H, Pakela J, Ten Haken RK, El Naqa I. Introduction to machine and deep learning for medical physicists. Med Phys. 2020;47(5):127–47.
Juba B, Le HS 2019 Precision-recall versus accuracy and the role of large data sets. In: proceedings of the AAAI conference on artificial intelligence, vol. 33, pp. 4039–4048
Junker M, Hoch R, Dengel A 1999 On the evaluation of document analysis components by recall, precision, and accuracy. In: proceedings of the fifth international conference on document analysis and recognition. ICDAR’99 (Cat. No. PR00318), pp. 713–716. IEEE
Powers DM 2020 Evaluation: from precision, recall and f-measure to roc, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061
Janizek JD, Celik S, Lee S-I. Explainable machine learning prediction of synergistic drug combinations for precision cancer medicine. BioRxiv. 2018;331769.
Khaled al-Bashiti M, Naser M. Verifying domain knowledge and theories on fire-induced spalling of concrete through explainable artificial intelligence. Constr Building Mater. 2022;348:128648.
Gramegna A, Giudici P. Shap and lime: an evaluation of discriminative power in credit risk. Front Artif Intell. 2021;4:752558. https://doi.org/10.3389/frai.2021.752558.
Javed U, Ijaz K, Jawad M, Ansari EA, Shabbir N, Kutt L, Husev O. Exploratory data analysis based short-term electrical load forecasting: a comprehensive analysis. Energies. 2021;14(17):5510.
Milo T, Somech A 2020 Automating exploratory data analysis via machine learning: An overview. In: Proceedings of the 2020 ACM SIGMOD International Conference on Management of Data, pp. 2617–2622
Acknowledgements
All authors are thanked for their contributions to this research, and on behalf of all authors, we would like to thank Anupam Kumar Bairagi for supervising and significantly contributing to this study.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
MBH contributed substantial conceptual and design contributions to the study. AS supported AD in assessing the data and appropriately drafting this report. Other writers contributed to the development of the manuscript’s preliminary draft. All authors reviewed the results and approved the final version of the paper.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to Participate
Not Applicable.
Consent for Publication
Not Applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hossain, M.B., Shama, A., Adhikary, A. et al. An Explainable Artificial Intelligence Framework for the Predictive Analysis of Hypo and Hyper Thyroidism Using Machine Learning Algorithms. Hum-Cent Intell Syst 3, 211–231 (2023). https://doi.org/10.1007/s44230-023-00027-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s44230-023-00027-1