Abstract
Chronic Kidney Disease (CKD) has become a major problem in modern times, and it is dubbed the silent assassin due to its delayed signs. To overcome these critical issues, early identification may minimize the prevalence of chronic diseases, though it is quite difficult because of different kinds of limitations in the dataset. The novelty of our study is that we extracted the best features from the dataset in order to provide the best classification models for diagnosing patients with chronic kidney disease. In our study, we used CKD patients’ clinical datasets to predict CKD using some popular machine learning algorithms. After handling missing values, K-means clustering has been performed. Then feature selection was done by applying the XGBoost feature selection algorithm. After selecting features from our dataset, we have used a variety of machine learning models to determine the best classification models, including Neural Network (NN), Random Forest (RF), Support Vector Machine (SVM), Random Tree (RT), and Bagging Tree Model (BTM). Accuracy, Sensitivity, Specificity, and Kappa values were used to evaluate model performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Chronic Kidney Disease (CKD) is a disorder in which the kidneys are damaged and unable to perform their vital function of filtering blood. As a consequence, excess fluid and waste from the blood persist in the body, which may lead to a variety of health concerns. Early on, there are no obvious signs of the disease, making it a silent killer. CKD occurs when a disease or condition compromises kidney function, resulting in kidney damage that worsens over time. Type 1 or type 2 diabetes [1], high blood pressure, glomerulonephritis, interstitial nephritis, prolonged blockage of the urinary system [2], vesicoureteral disorders [3], recurrent infection of the kidneys, and other such disorders may all lead to chronic kidney disease. It’s becoming more and more prevalent, and it’s largely accepted as a global problem. As a result of enhanced mortality and bleakness, a high risk of a number of different maladies, including heart disease and human services, has made it a long-term problem [4]. In the United States, an estimated 37 million individuals suffer from kidney disease (15 percent of the adult population; more than 1 in 7 adults). 90% of people with renal disease are unaware of their condition [5]. For CKD prediction, a number of studies have been undertaken, utilizing various machine learning models and determining their accuracy rates. Alaiad et al. [6] have presented a powerful prediction technique for early detection of chronic kidney disease. Their goal was to save patients’ lives while lowering treatment costs and human error hazards. The researchers used five algorithms, such as NB, J48, SVM, KNN, and JRip, to predict and diagnose CKD. Sobrinho et al. [7] have studied how machine learning tools may help to diagnose CKD early in developing nations. Based on their classification findings, the J48 decision tree was selected as the best machine learning approach after comparing it to other algorithms such as random forest, naive bayes, support vector machine, k-nearest neighbor, and multilayer perceptron. However, our study has attempted to analyze the value of features using feature engineering and aims to uncover the most essential features responsible for CKD. This study used the Chronic Kidney Disease Data Set [8], which comprises age, blood pressure, and a total of 25 relevant characteristics that have previously been used to classify patients with CKD. Classification has been accomplished via the use of a variety of supervised machine learning techniques [9], including NN, RF, SVM, RT, and BTM. Our main goal is to develop a prediction model that can offer a more accurate picture of CKD. The major contributions to this work are as follows:
-
We have used a clinical report dataset of key aspects of kidney patients’ for this research.
-
In order to minimize overfitting and underfitting, XGBoost has been employed to estimate the importance of features.
-
To test the model’s performance based on the datasets, two independent datasets are created: the whole dataset and the XGBoost dataset.
-
For the prediction of kidney disease, different machine learning algorithms have been used to train the datasets.
The flow of the paper is as follows: Section 2 includes a review of related research on Chronic Kidney Disease prediction using several classifiers. Section 3 shows the methodology of our literature, which includes data collection, data preprocessing, data clustering, feature selection, dataset splitting, and developing a model based on different machine learning algorithms. In Section 4, the result and discussion of our work have been demonstrated. Finally, the conclusion of our finding and our future planning have been discussed in Section 5.
2 Literature Review
V´asquez-Morales et al. [10] developed a Neural Network (NN)-based classifier to predict the probability of acquiring chronic kidney disease (CKD) in the Colombian population by taking two population categories into account. Using the test dataset, their model predicted the expected progression of the medical condition CKD. The precision of the forecast was supported by a case-based reasoning (CBR) example with an adequate explanation. As a test dataset for training and testing the proposed NN-CBR twin system model, the demographic data and medical care information of around 20,000 individuals with chronic kidney disease (CKD) and 20,000 individuals without CKD were employed. Using this dataset with more extensive characteristics, the suggested NN model with five layers accurately predicted the probability of getting CKD with a 95% precision. Sinha et al. [11] used the Support Vector Machine (SVM) and K-Nearest Neighbor (KNN) classifiers to predict Chronic Kidney Disease. The experimental findings show that the KNN classifier is more effective than the SVM classifier. SVM and KNN classifiers have an accuracy of 73.75 and 78.75 percent, respectively. Khan et al. [12] suggested machine learning (ML) techniques for the prediction of chronic kidney disease (CKD). The dataset extracted from the UCI ML pool had 400 occurrences. NBTree, J48, SVM, Logistic Regression, Multi-Layer Perceptron, and Naive Bayes were among the machine learning techniques used. The findings of the aforementioned methods were compared to the definition of the best accurate approach for identifying CKD and non-CKD patients. Exact experimental findings for accuracy were 95.75 percent for NB, 96.5 percent for LR, 97.2 percent for MLP, 97.75 percent for J48, 98.25 percent for SVM, and 98.75 percent with NBTree. Hosseinzadeh et al. [13] presented an Internet of Things-based diagnostic paradigm for chronic nephrosis. In their classification, they used Judgment Treaties (J48), SVM, MLP, and Naive Bayes classifiers. In comparison to support vector machines (SVM), multilayer perceptrons (MLP), and Naive Bayes classifiers, their experimental data demonstrated that the data collection implemented with their proposed model selection attained 97% precision, 99% sensitivity, and 95% specificity with the decision tree (J48) classifier. Gunarathne et al. [14] used machine learning classification methods to predict a patient’s CKD or non-CKD condition. To conduct their study, the researchers acquired a CKD dataset from the UCI repository that included 400 individual data records and 25 different features. Using a variety of machine learning techniques, such as Multiclass Decision Forest, Multiclass Decision Jungle, Multiclass Logistic Regression, and Multiclass Neural Network, the 14 selected characteristics for CKD patients from the reduced dataset were evaluated and predicted. Finally, the results of several models were compared, revealing that the model using the Multiclass Decision Forest algorithm performed the best, with an accuracy of 99.1% for the reduced dataset. Alasker et al. [15] found that the DT classifier can predict renal illness. The pool of UCI ML data was used for all of their studies and testing. They attained 98.41% accuracy with an error rate of 0.0159 when all 24 features of the dataset were employed, and 98.41% accuracy with an error rate of 0.0159 when just 8 characteristics were used. To categorize CKD, Abdullah et al. [16] assessed five feature selection techniques, including Random Forest feature selection, forward selection, forward exhaustive selection, backward selection, and backward exhaustive selection, as well as four machine learning algorithms. Implemented machine learning techniques included the Random Forest classifier, Linear and Radial SVM, Naive Bayes, and Logistic Regression. The performance of the machine learning algorithms was assessed in terms of accuracy, sensitivity, specificity, and AUC. The findings indicated that the Random Forest classifier with Random Forest feature selection was the best machine learning model for classifying CKD, with the greatest accuracy, sensitivity, specificity, and AUC (98.825%, 98.040%, 100%, and 98.9%, respectively). Charleonnan et al. [17] examined four machine learning methods, KNN, SVM, LR, and DT classifiers, to predict CKD in CKD patients. These models were developed using data from Indian patients of Apollo Hospitals, and their performance is being tested to establish the most accurate classifier for predicting chronic renal disease. In their experiments, they discovered that the SVM classifier had the highest accuracy, with a score of 97.3%. In addition, following training on the dataset, SVM had the highest sensitivity.
2.1 State of the Art of CKD Detection Models
Kidney Disease is a chronic disease that may lead to a range of health issues and a number of life difficulties. The disease exhibits no first signs, making it a silent killer. In recent decades, several researchers have utilized machine learning (ML) techniques in healthcare studies, particularly in the identification of kidney disease. Therefore, we included the listing of existing ML work in Table 1.
3 Methodology
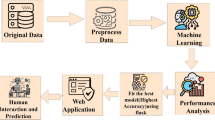
The proposed model is able to predict CKD patients at an early stage as a result of our usage of clinical features as a training dataset and the model’s capacity to predict CKD patients. To conduct this research, we have used the Chronic Kidney Disease Data Set [8]. After collecting the data, preprocessing the data is crucial to preparing it for modeling. Data preprocessing includes essential processes, including handling missing values, where we have used predictive mean matching (PMM) for handling missing values. PMM is an appealing approach, particularly when dealing with quantitative variables that do not follow a normal distribution. In order to locate the distinct groups within the collection of data, the dataset has been subjected to data clustering. We observed the ability of our classifiers using the initial features as well as the features selected by the feature selection process. The XGBoostbased feature selection technique was used to get the 7 most essential features from the 25 features. In building a predictive model, feature selection refers to assisting in extracting the best features, which has a great impact on the performance and execution times of the models. Using these initial and chosen features, two different datasets, the main dataset and the XGBoost dataset, were made for training purposes. After feature selection, building classification models requires splitting the data into training and testing sets, where we have assigned 80% of the data for the training phase and the remaining 20% for the testing phase. When constructing a model, the training set is used to develop the model, while the testing set is used to validate the model. Training data needs to be provided for the models to learn from it. Later, we applied different ML methods including NN, RF, SVM, RT, and BTM to look for useful patterns and translate the features of the input data. The models with classifiers are implemented to make a comparison with the other existing systems, and a different training model has been implemented for testing the dataset so that we can pick the best model for our reliable dataset. With a great accuracy of 100% on the XGBoost dataset, the process resulted in SVM being the most effective method. Furthermore, the diagnosis system has shown the most useful features of a patient affected by CKD. Following the utilization of different methods, we must describe the model performances in order to choose the best model through result analysis. Figure 1 depicts our model’s overall process and structure.
3.1 Data Description
In this study, the Chronic Kidney Disease Data Set from the UCI Machine Learning Repository has been used [8]. The dataset has 400 items and 25 characteristics that have been employed in this study. Age, blood pressure, specific gravity, albumin, and a total of 21 relevant attributes have been included in the dataset. Some of the features in this data set have continuous values. These include age, blood pressure, random blood glucose, blood urea, serum creatinine, sodium, potassium, hemoglobin, packed cell volume, white blood cell count, and red blood cell count. Additionally, there are binary values for several attributes, including specific gravity, albumin, sugar, red blood cells, pus cell, pus cell clumps, bacteria, hypertension, diabetes mellitus, coronary artery disease, appetite, pedal edema, and anemia.
3.2 Data Preprocessing
Throughout the dataset, missing values were recovered using predictive mean matching (PMM), and all variables were then categorized as having nominal values. Multiple imputation of missing data using PMM is an appealing approach, particularly when dealing with quantitative variables that do not follow a normal distribution. This is done by establishing a small subset of instances in which the result variable matches the outcome of cases with missing data. When compared to other imputation algorithms, it often produces less plausible results and better represents diverse data [18]. Figure 2 illustrates the dataset’s missing data plot.
According to the plot, we have found the highest missing value by wbcc of 26.50% and the lowest missing value by pe of 0.25%. The top three variables that have the highest missing value are wbcc, sod, and pc, respectively.
3.3 K-Means Clustering
Data clustering has been applied to the dataset in order to identify unique groups of data within the collection. K-means clustering was used with the dataset to achieve this objective. The optimum number of clusters, which was found to be three, was determined using the elbow approach. Three clusters, including the centroid point, were used in the K-means clustering. Figure 3 depicts the (a) Elbow Method, (b) K-means clustering, and (c) Dendrogram of clustering.
3.4 XGBoost Based Feature Selection
Feature selection reduces the number of input variables needed to develop a predictive model. Decreasing the number of input variables may both lower computing costs and improve model accuracy [19]. The XGBoost approach with SHAP (Shapley Additive Explanations) value analysis has been used in this case to choose the features. XGBoost is a decision tree-based gradient boosting ensemble machine learning technique. We use gradient boosting for regression and classification. It works by calculating weak classifiers iteratively [20]. Using XGBoost, an effective feature selection has been made to increase the performance of models. By removing features that are unneeded, redundant, or irrelevant to the activity at hand, a good feature selection approach can improve the effectiveness of machine learning algorithms. A redundant attribute is one that has a strong association with other characteristics while also lowering accuracy. The most important features were extracted using a method of feature selection. From a global perspective, an essential trait that is closer to the root node has a greater relevance value. To generate relevance scores for features, several boosting tree techniques, such as the XGBoost, can be used [21]. The top reasons for the feature importance rankings of XGBoost are gain, cover, and frequency [22]. The averaged gain throughout all splits where this feature has been used is called Gain, whereas the average coverage across all splits is called Cover [23]. In this study, the word benefit has been used to assess feature importance. The importance of each attribute was first determined using the XGBoost method, and then the results were compared. Each feature is given a weighted significance value by the XGBoost algorithm. All of the attributes are ranked [24] according to their importance.
We determined the most feasible feature subset based on these rating factors.
3.5 Dataset Splitting
We have separated our data set into a training set and a testing set. For the training set we have allocated 80% of the total dataset, and for the testing set, 20% of the dataset has been allocated. We used R’s trainControl package in conjunction with the cross-validation technique on the test dataset. The proposed model is tested and validated using a total of 20% of the dataset. Using this method, we were able to significantly cut down on the model’s overfitting problem.
3.6 Applying Machine Learning Algorithms
3.6.1 Neural Network (NN)
NN is a collection of neurons that receive input and generate output using information from other nodes. They basically tackle issues through trial and error. One of the most powerful aspects of a neural network is the way basic neurons are coupled to produce a complex system. Each neuron can make basic mathematical judgments. Many neurons working together can solve complicated problems accurately. A shallow network has three layers: input, hidden, and output. A deep neural network includes several hidden layers, increasing the complexity of the issues it can solve [25].
Almansour et al. [26] have used machine learning to detect CKD early in the course of the disease. This study used the Artificial Neural Network (ANN) and Support Vector Machine (SVM) for classification. The final models for both proposed techniques used the best-available parameters and features, and their experiments showed that ANN outperformed SVM, with accuracy rates of 99.75%.
3.6.2 Random Forest (RF)
RF is a technique that builds on bagged trees. Several benchmarking tests have shown that it is one of the finest machine learning approaches presently available for classification issues. RF is a statistical approach based on the analysis of data. While the statistical model is important, iteratively training the algorithm is more important than the method’s mathematical formulation [27]
Hore et al. [28] have suggested a genetic algorithm-trained neural network (NNGA) for the detection of chronic kidney disease (CKD). On the other hand, the model was compared to Neural Network, Multilayer Perceptron Feedforward Network and Random Forest. With 92.54% accuracy, 85.71% precision, 96% recall, and 90.56% F-Measure, Random Forest performed modestly.
3.6.3 Support Vector Machine (SVM)
SVM is a technique for supervised machine learning that may be used for classification and regression [29]. It uses a linear discriminant function to take a small number of meaningful limit samples from each class and separate them as widely as feasible. Nonlinear function components may be added to these systems, enabling them to create quadratic, cubic, and higher-order decision limits beyond the linear restrictions.
Khan et al. [12] have employed three alternative models, and the SVM model, which addressed noise disturbance in the composite dataset, attained 98.25% accuracy in this research for predicting CKD. One of the most accurate models was this one, but it also had the greatest scores in other assessment parameters, such as precision (98.30%) and F-measure (98.30%).
3.6.4 Random Tree (RT)
RT is an ensemble learning method and supervised classifier that is trained using examples. Moreover, it is able to handle both classification and regression issues [30]. A random selection of characteristics is utilized for each split, and therefore, it’s similar to a decision tree in operation. The term”forest” refers to a collection of random trees. This classifier uses the input feature set to classify the input for each individual tree in the forest. The random tree’s output chooses from the most popular votes [31].
Almustafa et al. [32] have used several classifiers to classify a CKD dataset. The J48 and decision table classifiers beat the other classifiers, where the Random Tree classifier achieved 95.50% accuracy.
3.6.5 Bagging Tree Model (BTM)
BTM is an ensemble approach, which combines numerous decision trees to generate higher predicting performance than using a single tree. When a collection of weak learners comes together, they become stronger, and this is the underlying premise behind the ensemble model. As opposed to one decision tree, bagged trees rely on a large number of decision trees, making it possible to draw on the wisdom of a variety of different models [33].
Hasan et al. [34] have suggested an ensemble approach-based classifier to enhance the classification of CKD. The system’s performance is also examined using tenfold cross-validation and the receiver operating characteristic curve. Extensive testing on CKD datasets revealed that the BTM model has obtained. 96.00% classification accuracy.
4 Results and Discussion
4.1 Feature Importance Finding
Feature significance approaches are those that produce a score for each of a model’s input attributes; the scores just indicate the”importance” of each feature [35]. A higher score suggests that the specific attribute will have a bigger influence on the model used to forecast a certain variable [36]. Figure 4 shows SHAP values for the dataset’s features, where feature names are listed on the y-axis in order of importance from top to bottom, and SHAP values for the corresponding features are listed on the x-axis.
In the dataset that has been used in our literature, “hemo” has a distribution of values between -0.075 and 0.10, which is considered the widest distribution, and so “hemo” has a high impact on CKD. Within SHAP value, “sg” was the second most common cause of CKD among all features. “pcv” is the third cause of CKD according to its SHAP value distribution. The SHAP value distribution of “bp” is less than all other features, so it is considered the least impactful feature on CKD.
The Gain, Cover, Frequency, and Importance values obtained using the XGBoost technique play a crucial role in finding the dataset’s key features. The feature “hemo” has gained the highest place in Table 2 in terms of these four values. Gain, Cover, and Frequency values for “hemo” are 0.7753, 0.5126, and 0.3148 respectively. In term of “sg”, it holds the second best position and the values of Gain, Cover, and Frequency for this feature are 0.1848, 0.1533, and 0.1852. In the rest of all features, “bp” has the poorest Gain and frequency values, and although it has a higher Cover value than “sc”, it has not been considered for our study. In our literature, we have focused on six attributes out of seven attributes based on the values of Gain, Cover, and Frequency “hemo”, “sg”, “pcv”, “al”, “pc”, and “sc”. Table 2 shows the values of Gain, Cover, and Frequency for the dataset’s features.
In the above Fig. 5, in term of feature “hemo”, when the SHAP value is between -0.05 to 0.00 and 0.05 to 0.10, it is more scattered between 5 to 15. “sg”is more scattered between 1 to 3 when SHAP value is between -0.04 to 0.00. The SHAP value of “pcv”is 0.00 then it is more scattered between 10 to 45. In term of “al”, when the SHAP value is between − 0.05 to 0.00, it is more scattered between 1.5 and 5. For the feature “pe”, when SHAP value is between − 0.01 to 0.00, it is more distributed at 0.00. In terms of “bp”, it is more scattered between 80 and 120, when the SHAP value is between − 0.0005 and 0.000.
4.2 Comparison of Various Algorithms on the Different Features
The performance of all models on the full dataset and also on the XGBoost dataset is shown in Table 3.
From the above Table 3, it is noticed that the three of our applied models, NN, RF, and SVM, have provided the best performance in terms of accuracy, sensitivity, specificity, and kappa on the full dataset, whereas the performance of SVM is highest on the XGBoost dataset. On the full dataset, NN delivered the best results. The poorest accuracy (96.25%) has been given by both RT and BTM on the full dataset, and they have also provided the lowest sensitivity (94.34%) on the full dataset. The kappa value of RT was 91.84% on both the full dataset and the XGBoost dataset, which was the worst. Hence, it is needless to mention that the overall performance of all the applied models has enhanced after XGBoost feature selection.
4.2.1 Comparison Between Different Methods Based on Accuracy
Accuracy is one of the most important performance metrics to evaluate machine learning algorithms. As mentioned earlier, we have used five classifiers. We have applied the five different methods to the original 25 input features and then to the 7 input features selected by the XGBoost approach. Figure 6 shows the accuracy of various kinds of classifiers.
Considering 25 features, the NN Classifier obtained the best accuracy of 100%, whereas the accuracy of RT and BTM (96.25%) was the lowest. The accuracy of RF and SVM was similar to each other (98.75%). Evaluating only the 7 selected XGBoost features, the RT Classifier generated the lowest accuracy (96.25%) while the SVM had an outstanding performance of 100% accuracy. We got 97.5%, 98.75%, 97.5% accuracy for NN, RF, and BTM, respectively, with the 7 XGBoost features.
4.2.2 Comparison Between Different Methods Based on Sensitivity
Sensitivity score is a vital performance metrics to classify the people with chronic kidney disease accurately. Figure 7 shows the sensitivity scores for the various algorithms and feature sets.
Using the original 25 features, the RT and BTM algorithms generated the lowest sensitivity score (94.34%), whereas the NN algorithm achieved the highest sensitivity score (100%). RF and SVM had similar sensitivity scores of 98.04% based on the 25 features. For the 7 selected XGBoost features, the RT algorithm had the lowest sensitivity score (94.34%) while BTM, NN, and RF provided better results. BTM, NN, and RF algorithms got 96.15%, 98% and 98.04% respectively. The best sensitivity score (100%) was obtained with SVM applied to the 7 XGBoost features.
4.2.3 Comparison Between Different Methods Based on Specificity
Another important performance metric for accurately classifying people with chronic kidney disease is the specificity score. Figure 8 shows the specificity scores for the various classification algorithms and feature sets.
The applied models, NN, RF, SVM, RT, and BTM, have provided the best specificity score (100%) using the original 25 features. The lowest specificity score (96.67%) was generated by the NN algorithm, whereas the other four algorithms generated the best specificity score (100%) applying the 7 selected XGBoost features.
4.2.4 Comparison Between Different Methods Based on Kappa
Kappa score is considered a good performance metric to accurately classify people with chronic kidney disease. Figure 9 shows the Kappa scores for the various classification algorithms and feature sets.
Using the original 25 features, the RT and BTM algorithms generated the lowest kappa score (91.84%), whereas the NN algorithm achieved the highest kappa score (100%). RF and SVM had similar kappa scores of 97.32% based on the 25 features. The RT algorithm had the lowest kappa score (91.84%) while BTM, NN, and RF obtained better outcomes considering the 7 selected XGBoost features. BTM, NN, and RF algorithms got 94.59%, 94.67% and 97.32% respectively. The best kappa score (100%) was obtained with SVM applied to the 7 XGBoost features.
4.3 Comparative Evaluation of Our Proposed Model
To conduct our study, we used five different models to evaluate our model’s accuracy to that of existing CKD prediction systems. Table 4 highlights our model’s overall performance in comparison to other systems.
Table 4 provides an overall view of the performance of the algorithms in our investigation when compared to previous studies. Experimenting with all characteristics yielded the highest accuracy (100%) with the NN model, while a lower accuracy of 96.25% was attained with both the RT and BTM models. Existing studies show that the NN model has obtained 99.75% accuracy, while the BTM and RT models have achieved 96.00% and 95.50% accuracy, respectively. For the RF, SVM, and RT models, we have gained 98.75% accuracy for both the RF and SVM models and 96.25% accuracy for the RT model. In this case, existing work has reached 92.54%, 98.25%, and 95.50% accuracy via RF, SVM, and RT models, respectively. Each row of the table represents an algorithm that was utilized in our investigations, as well as related studies and the findings that were presented. When compared to other studies, the results of our proposed models seem to be pretty good.
5 Conclusion
The CKD diagnosis is a difficult challenge. In our literature, we have presented a predictive model using different machine learning algorithms, including NN, RF, SVM, RT, and BTM, to predict CKD earlier. We mainly focus on the empirical comparisons of those mentioned ML algorithms. Based on the empirical results of the applied algorithms, the NN, RF, and SVM have given the highest accuracy on the full dataset. Moreover, comparing overall accuracy metrics, NN has given the best performance on the full dataset, and SVM has given the highest performance on the XGBoost dataset. Our study has limitations due to the small size of the dataset used. In the future, we will use our developed models on other datasets of other diseases and try to develop more advanced and expert systems.
Data Availability
This research study is based on an open source dataset. Dataset can accessed from this link: https://archive.ics.uci.edu/ml/datasets/Chronic Kidney Disease.
References
Ohta M, Babazono T, Uchigata Y, Iwamoto Y. Comparison of the prevalence of chronic kidney disease in Japanese patients with type 1 and type 2 diabetes. Diabet Med. 2010;27(9):1017–23.
Dimitrijevic Z, Paunovic G, Tasic D, Mitic B, Basic D. Risk factors for urosepsis in chronic kidney disease patients with urinary tract infections. Sci Rep. 2021;11(1):1–8.
van der Plas E, Lullmann O, Hopkins L, Schultz JL, Nopoulos PC, Harshman LA. Associations between neurofilament light-chain protein, brain structure, and chronic kidney disease. Pediatric Res. 2021;91:135–40.
Couser WG, Remuzzi G, Mendis S, Tonelli M. The contribution of chronic kidney disease to the global burden of major noncommunicable diseases. Kidney Int. 2011;80(12):1258–70.
Phillips S, Knuchel N. Chronic kidney disease: nutrition basics. J Ren Nutr. 2011;21(4):15–7.
Alaiad A, Najadat H, Mohsen B, Balhaf K. Classification and association rule mining technique for predicting chronic kidney disease. J Inf Knowl Manag. 2020;19(01):2040015.
Sobrinho A, Queiroz ACDS, Da Silva LD, Costa EDB, Pinheiro ME, Perkusich A. Computer-aided diagnosis of chronic kidney disease in developing countries: A comparative analysis of machine learning techniques. IEEE Access. 2020;8:25407–19.
Avci E, Karakus S, Ozmen O, Avci D. Performance comparison of some classifiers on chronic kidney disease data. In: 2018 6th international symposium on digital forensic and security (ISDFS). IEEE; 2018. p. 1–4.
Hassan MM, Mollick S, Yasmin F. An unsupervised cluster-based feature grouping model for early diabetes detection. Healthcare Anal. 2022;2:100112.
V’asquez-Morales GR, Martinez-Monterrubio SM, Moreno-Ger P, Recio-Garcia JA. Explainable prediction of chronic renal disease in the colombian population using neural networks and case-based reasoning. IEEE Access. 2019;7:152900–10.
Sinha P, Sinha P. Comparative study of chronic kidney disease prediction using knn and svm. Int J Eng Res Technol. 2015;4:608–12.
Khan B, Naseem R, Muhammad F, Abbas G, Kim S. An empirical evaluation of machine learning techniques for chronic kidney disease prophecy. IEEE Access. 2020;8:55012–22.
Hosseinzadeh M, Koohpayehzadeh J, Bali AO, Asghari P, Souri A, Mazaherinezhad A, Bohlouli M, Rawassizadeh R. A diagnostic prediction model for chronic kidney disease in internet of things platform. Multimedia Tool Appl. 2021;80(11):16933–50.
Gunarathne WHSD, Perera KDM, Kahandawaarachchi KADCP. Performance evaluation on machine learning classification techniques for disease classification and forecasting through data analytics for chronic kidney disease (ckd). In: 2017 IEEE 17th international conference on bioinformatics and bioengineering (BIBE). IEEE: UK; 2017. p. 291–6.
Alasker H, Alharkan S, Alharkan W, Zaki A, Riza LS. Detection of kidney disease using various intelligent classifiers. In: 2017 3rd international conference on science in information technology (ICSITech). IEEE; 2017. p. 681–4.
Abdullah AA, Hafidz SA, Khairunizam W. Performance comparison of machine learning algorithms for classification of chronic kidney disease (CKD). J Phys: Conf Ser. 2020;1529(5):052077.
Charleonnan A, Fufaung T, Niyomwong T, Chokchueypattanakit W, Suwannawach S, Ninchawee N. Predictive analytics for chronic kidney disease using machine learning techniques. In: 2016 management and innovation technology international conference (MITicon). IEEE: UK; 2016. p. 80–3.
Austin PC, White IR, Lee DS, van Buuren S. Missing data in clinical research: a tutorial on multiple imputation. Can J Cardiol. 2021;37(9):1322–31.
Hassan MM, Khan MAR, Islam KK, Hassan MM, Rabbi MMF. Depression detection system with statistical analysis and data mining approaches. In: 2021 international conference on science & contemporary technologies (ICSCT). IEEE; 2021. p. 1–6.
Wang D, Zhang Y, Zhao Y (2017) Lightgbm: an effective mirna classification method in breast cancer patients. In: Proceedings of the 2017 International Conference on Computational Biology and Bioinformatics, pp. 7–11
Manju N, Harish B, Prajwal V. Ensemble feature selection and classification of internet traffic using xgboost classifier. Int J Comp Netw Informat Secur. 2019;10(7):37.
Guo J, Yang L, Bie R, Yu J, Gao Y, Shen Y, Kos A. An xgboost-based physical fitness evaluation model using advanced feature selection and bayesian hyper-parameter optimization for wearable running monitoring. Comput Netw. 2019;151:166–80.
Chakraborty S, Bhattacharya S. Application of xgboost algorithm as a predictive tool in a cnc turning process. Rep Mechan Eng. 2021;2(1):190–201.
Ghosh P, Azam S, Jonkman M, Karim A, Shamrat FJM, Ignatious E, Shultana S, Beeravolu AR, De Boer F. Efficient prediction of cardiovascular disease using machine learning algorithms with relief and lasso feature selection techniques. IEEE Access. 2021;9:19304–26.
Kunwar, V., Chandel, K., Sabitha, A.S., Bansal, A. (2016) Chronic kidney disease analysis using data mining classification techniques. In: 2016 6th International Conference–Cloud System and Big Data Engineering (Confluence), pp. 300–305
Almansour NA, Syed HF, Khayat NR, Altheeb RK, Juri RE, Alhiyafi J, Alrashed S, Olatunji SO. Neural network and support vector machine for the prediction of chronic kidney disease: a comparative study. Comput Biol Med. 2019;109:101–11.
Subasi A, Alickovic E, Kevric J. Diagnosis of chronic kidney disease by using random forest. In: Badnjevic A, editor. CMBEBIH 2017. Singapore: Springer; 2017. p. 589–94.
Ceyhan M, Orhan Z, Domnori E. Health service quality measurement from patient reviewsin turkish by opinion mining. In: Badnjevic A, editor. CMBEBIH 2017. Singapore: Springer; 2017. p. 649–53.
Hassan MM, Hassan MM, Akter L, Rahman MM, Zaman S, Hasib KM, Jahan N, Smrity RN, Farhana J, Raihan M, et al. Efficient prediction of water quality index (wqi) using machine learning algorithms. Human-Centric Intell Sys. 2021;1(3–4):86–97.
Basar MD, Aydın A. Chronic kidney disease prediction with reduced individual classifiers. Electrica. 2018;18(2):249–55.
Chittora P, Chaurasia S, Chakrabarti P, Kumawat G, Chakrabarti T, Leonowicz Z, Jasinski M, Jasin`ski L, Gono R, Jasin`ska E, et al. Prediction of chronic kidney disease-a machine learning perspective. IEEE Access. 2021;9:17312–34.
Almustafa KM. Prediction of chronic kidney disease using different classification algorithms. Inform Med Unlock. 2021;24:100631.
Wang W, Chakraborty G, Chakraborty B. Predicting the risk of chronic kidney disease (ckd) using machine learning algorithm. Appl Sci. 2020;11(1):202.
Zubair Hasan KM, Zahid Hasan M. Performance evaluation of ensemble-based machine learning techniques for prediction of chronic kidney disease. In: Shetty NR, Patnaik LM, Nagaraj HC, Hamsavath PN, Nalini N, editors. Emerging research in computing, information, communication and applications. Singapore: Springer; 2019. p. 415–26.
Altmann A, Tolosi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26(10):1340–7.
Li C, Zhang Z, Ren Y, Nie H, Lei Y, Qiu H, Xu Z, Pu X. Machine learning based early mortality prediction in the emergency department. Int J Med Informatics. 2021;155:104570.
Acknowledgements
All authors are thanked for their contributions to this research, and on behalf of all authors, we would like to thank Md. Mehedi Hassan for supervising and significantly contributing to this study.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
MMH contributed substantial conceptual and design contributions to the study. MMH supported MMH in assessing the data and appropriately drafting this report. Other writers contributed to the development of the manuscript’s preliminary draft. All authors reviewed the results and approved the final version of the paper.
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare they have no conflicts of interest.
Ethics Approval
Not Applicable.
Consent to Participate
Not Applicable.
Consent for Publication
Not Applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hassan, M.M., Hassan, M.M., Mollick, S. et al. A Comparative Study, Prediction and Development of Chronic Kidney Disease Using Machine Learning on Patients Clinical Records. Hum-Cent Intell Syst 3, 92–104 (2023). https://doi.org/10.1007/s44230-023-00017-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s44230-023-00017-3