Abstract
Aiming at the utilization of adjacent image correlation information in multi-target segmentation of 3D image slices and the optimization of segmentation results, a 3D grouped fully convolutional network fused with conditional random fields (3D-GFCN) is proposed. The model takes fully convolutional network (FCN) as the image segmentation infrastructure, and fully connected conditional random field (FCCRF) as the post-processing tool. It expands the 2D convolution into 3D operations, and uses a shortcut-connection structure to achieve feature fusion of different levels and scales, to realizes the fine-segmentation of 3D image slices. 3D-GFCN uses 3D convolution kernel to correlate the information of 3D image adjacent slices, uses the context correlation and probability exploration mechanism of FCCRF to optimize the segmentation results, and uses the grouped convolution to reduce the model parameters. The dice loss that can ignore the influence of background pixels is used as the training objective function to reduce the influence of the imbalance quantity between background pixels and target pixels. The model can automatically study and focus on target structures of different shapes and sizes in the image, highlight the salient features useful for specific tasks. In the mechanism, it can improve the shortcomings and limitations of the existing image segmentation algorithms, such as insignificant morphological features of the target image, weak correlation of spatial information and discontinuous segmentation results, and improve the accuracy of multi-target segmentation results and learning efficiency. Take abdominal abnormal tissue detection and multi-target segmentation based on 3D computer tomography (CT) images as verification experiments. In the case of small-scale and unbalanced data set, the average Dice coefficient is 88.8%, the Class Pixel Accuracy is 95.3%, and Intersection of Union is 87.8%. Compared with other methods, the performance evaluation index and segmentation accuracy are significantly improved. It shows that the proposed method has good applicability for solving typical multi-target image segmentation problems, such as boundary overlap, offset deformation and low contrast.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
2D and 3D image segmentation, and video target detection and tracking are currently hot issues in the field of image analysis and deep learning. The principle of image multi-target instance segmentation is pixelwise classification for multi-objects with semantic labels, that is, using a set of object categories to classify all the pixels of the image, to segment and describe multi-objects of interest in the image [1]. With the development of image analysis theory and deep learning, a variety of effective multi-target image segmentation models and algorithms are proposed [2,3,4,5]. However, due to the influence of noise, offset deformation, gray value distortion and local position effect in some complex scenes, the existing methods still have problems in multi-target segmentation, such as missing detection targets, segmentation position offset, unclear boundary and so on. Especially in 3D image segmentation, there is information association between adjacent 2D slices. There are still many bottlenecks to be solved in using the spatiotemporal association information between slices to guide and constrain the detection and segmentation of current image slices [6, 7]. At the same time, some methods have good performance in single-target segmentation, but there are still some challenging problems to be solved for the multi-target instances segmentation of complex images [8, 9].
At present, traditional methods and deep learning are mainly used for image multi-target detection and segmentation. Traditional algorithms mostly use gray-scale features to segment [10]. Classical methods include threshold-based [11], edge-based [12], and region-based image segmentation [13]. For the threshold-based image segmentation algorithm, when the segmentation target and the background differ greatly, the segmentation result is significant, but it is greatly affected by the contrast of gray features. Edge-based image segmentation algorithm mainly detects edge pixels, and then connects the detected points to form a target contour to achieve segmentation. The advantage is that it can suppress noise and keep the segmentation boundary clear, but it is less effective for low-resolution and blurred images. Region-based image segmentation algorithm uses the similarity of grayscale, color and texture between pixels to achieve segmentation. Its advantage is that it can extract regional features better, but it is easy to over-segment the image. In summary, traditional segmentation methods are greatly affected by subjective factors, and the preprocessing process is complicated. For example, the selection of seed points during segmentation and the thresholds of the segmentation boundary which require strong prior knowledge. At the same time, the selection of key parameters also has an important impact on segmentation accuracy. These problems make the traditional algorithms have many limitations to be applied in complex image semantic segmentation.
Deep convolutional neural network (DCNN) is an effective method for image feature extraction and analysis, and has been widely used in image classification, image generation, and target detection. Many excellent models have been proposed, such as AlexNet [14], ResNet [15], Faster R-CNN [16], etc. Although DCNN has strong feature extraction ability, when applied to complex image segmentation, it is easy to be affected by noise, image clarity, saturation, etc., as well as the complex scene target distribution structure and content diversity of the image itself, which increase the difficulty of image multi-target feature recognition, detection and segmentation [17, 18].
Multi-target segmentation of 3D image slices and short video data is currently a hot research topic. The deep learning models represented by the 3D convolutional neural network (3D CNN) provide powerful methods and tools for solving the above-mentioned problems. 3D CNN and its various variants [19,20,21,22] are essentially an extension of the 2D convolution model, which can provide a method for representation and learning of 3D image and video data. However, the computational complexity of 3D CNN increases exponentially compared to 2D networks, which makes 3D convolutions currently mainly used for feature learning based on slices or clips (clip-based). Usually, several slices are selected from the 3D data volume, or a short segment (such as 8 frames) is randomly sampled in the video for representation learning. For example, magnetic resonance imaging (MRI) and computer tomography (CT) in the medical field are all 3D image data volumes. When performing medical diagnosis and analysis, 3D image data is generally superimposed frame by frame on the time axis by 2D image slices. Therefore, extending the 2D convolution operation to the time domain can construct a 3D convolution operator that simultaneously processes both spatial and temporal information. By using the 3D convolution kernel to represent and calculate the associated information of adjacent slices, and taking the weighted average of the prediction of all slices, the segmentation result of the target slice can be obtained. The 3D convolution block can be easily integrated into existing 2D CNNs to achieve target segmentation of 3D image slices. In video image target detection, by designing and adopting the method for scale drawing calculating depending on the line detector and length detector, the image is easy to read and target detection is realized based on shape representation and contour features [23]. In software development effort estimation, a novel adaptive neuro-fuzzy inference system with a hybrid learning algorithm was been proposed [24]. The mean magnitude relative-error and the coefficient of correlation were used as assessment indices. This method can also be used to evaluate the segmentation results of target image.
Fully connected condition random field (FCCRF) is a probability distribution model proposed by Lafferty et al. [24] to solve the problem of sequence labeling. It is represented by an undirected graph, and all random variables are considered to follow the Markov properties. For image segmentation, FCCRF can be used as a post-processing tool for target segmentation, and its inherent context correlation and probability exploration mechanism can be used to optimize the segmentation results. Especially in the case of unclear segmentation boundary and poor continuity, the segmentation optimization effect of FCCRF is particularly significant.
If the short-term spatio-temporal feature modeling and information association mechanism of 3D convolution kernel, as well as the optimization ability of FCCRF for segmentation results can be utilized, and the grouped convolution strategy is adopted, it can provide an effective method for 3D image slice target segmentation, reduce the complexity of the algorithm, and realize the optimization of segmentation results.
In this paper, aiming at the utilization of adjacent slice related information in the multi-target segmentation of 3D image slices and the optimization of segmentation results, a 3D fully convolutional neural network segmentation model with grouped convolutional structure and FCCRF as a post-processing tool is proposed. It uses a FCN for image segmentation, uses a 3D convolutional kernel operation to extract the correlation information between individual slices, and determines the segmentation result of the target slice by weighted averaging the prediction results of adjacent slices. In the post-processing with FCCRF, its internal mechanism of the combination context correlation and probability exploration is used to optimize the segmentation results, and it is embedded between the last \(1\times 1\) convolutional layer and the output layer of the network, as an optimization step for the segmentation results of the basic FCN model. FCCRF receipts the original image and the basic FCN’s prediction segmentation image as inputs, and optimizes the pixel classification result by calculating the Gaussian penalty function between pixels assigned with different labels, and finally obtains a more refined prediction segmentation image, as well as continuous and clear segmentation boundary. At the same time, aiming at the problems of high computational complexity and time-consuming training of 3D convolution operation, the grouped convolution is applied to 3D FCN to construct a 3D grouped fully convolutional neural network model.
In the mechanism, the proposed method can comprehensively realize the correlation of information between adjacent slices and image pixels of 3D image, the feature fusion of different levels and scales, and the optimization of segmentation results, so as to reduce the algorithm complexity of 3D FCN and the influence of background pixels on target image segmentation. It has good applicability for complex image fine segmentation with problems such as insignificant morphological features of target image, weak correlation of spatial information and discontinuous target segmentation results.
Using the 3DIRCADB data set [25], the liver region segmentation of the abdominal 3D computer tomography (CT) image is used as an experiment to verify the effectiveness of the model and algorithm.
The novelty and main work of this paper are as follows:
-
(1)
A novel grouped 3D fully convolutional neural network multi-target image segmentation model fused with fully connected conditional random field is proposed. In mechanism, the 3D convolution operation can be used to represent and learn the associated information of adjacent slices, as well as the internal context information association and probability exploration mechanism of FCCRF, which can effectively improve the accuracy of complex 3D image slices multi-target instances segmentation.
-
(2)
The 3D-GFCN image segmentation module associates the segmentation target slice with the information of the adjacent corresponding segmentation area to reduce the influence of noise, offset distortion, and gray value distortion on the segmentation result. At the same time, a shortcut-connection structure is adopted to add low-level and high-level feature maps to achieve feature fusion and improve the accuracy of the segmentation results.
-
(3)
In this paper, FCCRF is used as the segmentation post-processing tool of 3D-GFCN to refine and optimize the segmentation results. The grouped convolution structure is used to greatly reduce model parameters and the complexity of 3D convolution operations. At the same time, in the algorithm design, the dice loss function which can reduce the influence of background pixels is used as the training objective function to effectively improve the imbalance between the number of background pixels and target pixels.
In the introduction, the current challenges in the multi-target segmentation of complex 3D images, as well as the current research status of image segmentation methods and technologies are reviewed and analyzed, and the ideas and algorithm strategy of a novel 3D-GFCN segmentation model established in this paper are pointed out. In Sect. 2, the 3D-GFCN model is established and its theoretical properties are analyzed. In Sect. 3, the comprehensive learning algorithm of 3D-GFCN is proposed. In Sect. 4, the multi-target segmentation experiment and result analysis for 3D medical image slices are carried out. Finally, the work of this paper is summarized, and the advantages and limitations of the method are pointed out.
2 3D Grouped Fully Convolutional Network Model
Aiming at the multi-target semantic segmentation of 3D image slices, in this section, a 3D grouped fully convolutional network model (3D-GFCN) segmentation model based on residual structure, which using fully connected conditional random fields as post-processing tools is proposed. The model takes the FCN as the basic structure, and by extending the existing 2D convolution to 3D convolution operation, improves the feature extraction method of traditional FCN using shortcut-connection and grouped convolution, and uses FCCRF to optimization segment the target pixels, so as to realize the image multi-target fine segmentation.
2.1 Fully Convolutional Network Segmentation Model
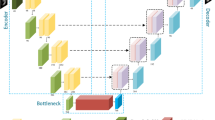
The 2D fully convolutional network is an important basic model in the field of image segmentation, which includes fully convolutional down-sampling feature extraction, deconvolution up-sampling target region detection, target semantic segmentation units, and shortcut-connection structure. The basic structure of FCN is shown in Fig. 1.
The shortcut-connection structure of FCN is the key to realize fine segmentation. The low-level feature maps of the image contain rich detailed information, while the high-level feature maps contain clearer semantic information. By adding and fusing them together, more accurate feature representation and semantic segmentation of the target region can be achieved. The shortcut-connection structure and information processing flow of FCN are shown in Fig. 2.
2.2 3D Grouped Fully Convolutional Network
In this section, by defining the 3D convolution operation and adopting the grouped convolution strategy, and taking the fully connected conditional random field as the post-processing tool, the 3D grouped convolution structure is built, and the 3D grouped fully convolution network fused with conditional random fields and end-to-end information processing flow are established.
2.2.1 3D Convolution Operation
The objects processed by 3D convolution mainly include 3D images and short video data, for example, 3D MRI and CT in the medical field. The 3D image data can be regarded as the frame-by-frame superposition of the 2D slice on the time axis. Therefore, the 2D convolution operation can be extended to the time domain to construct a 3D convolution operator which can process both spatial and temporal information, to realize the information association between adjacent slices of 3D image and short video frame data. The mathematical expression of the 3D convolution operation is as follows:
where \(t_{ij}^{x,y,z}\) represents the feature value at the (x, y, z) point of the \(j\mathrm{th}\) feature map in \(i\mathrm{th}\) layer, and \(\sigma \) is the activation function. M is the number of feature maps in \(i\mathrm{th}\) layer, and \((P_i,Q_i,R_i)\) is the width, height and depth of the 3D convolution kernel in \(i\mathrm{th}\) layer, respectively. (p, q, r) is the point coordinates of the 3D convolution kernel, and \(W_{ijm}^{pqr}\) represents the weight at (p, q, r) in the \(m\mathrm{th}\) convolution kernel of the \(j\mathrm{th}\) feature map in \(i\mathrm{th}\) layer. \(t_{(i-1)m}^{(x_0+p)(y_0+q)(z_0+r)}\) represents the value at \((x_0+p, y_0+q, z_0+r)\) of the \(m\mathrm{th}\) feature map in \((i-1)\mathrm{th}\) layer, and \((x_0,y_0,z_0)\) represents the left vertex coordinate in the (P, Q, R) neighborhood with (x, y, z) as the center point. \(b_{ij}\) represents the offset of the \(j\mathrm{th}\) feature map in \(i\mathrm{th}\) layer.
The 3D convolution kernel and 3D convolution operation defined by Eq. (1) have strong short-term spatiotemporal feature representation and modeling ability. The 3D CNN based on it can be used as an effective method to establish the relationship between adjacent slice features of the 3D image.
2.2.2 3D Grouped Convolution Structure
The grouped convolution groups the number of input feature map channels to reduce the amount of convolution kernel parameters. In this paper, the grouped convolution strategy is applied to 3D FCN. In the classic grouped convolution method [26], each channel of the input feature map is divided into g groups, and the channels between the groups are not connected, causing information loss and reducing the feature extraction and representation capabilities of the network. In this paper, the traditional grouped convolution method is improved, and the interleaving splicing is adopted to enhance the information association between each grouped channel. The \(3\times 3\times 3\) 3D volume integration grouped staggered splicing structure is shown in Fig. 3.
In Fig. 3, g is the number of groups, \(C_\mathrm{in}\) is the number of channels in the input feature map, and \(C_\mathrm{out}\) is the number of channels in the output feature map. It can be seen that the number of convolution kernel parameters of grouped convolution is 1/g of conventional convolution. In the case of a deep network, the network training parameters can be greatly reduced.
2.2.3 3D Grouped Fully Convolutional Network Segmentation Model
Aiming at the multi-target segmentation of 3D which composed of image slices, in this paper, a 3D grouped fully convolutional neural network model (3D-GFCN) is proposed. The model takes FCN as the basic segmentation model, uses a grouped interleaved splicing convolution structure to perform convolution operations, and uses a FCCRF as a post-processing tool to optimize the segmentation results. The specific structure of the model is shown in Fig. 4.
In Fig. 4, 3D-GFCN contains 19 convolution layers, 3 pooling layers, 3 up-sampling layers, as well as cropping and filling operations that make different feature maps have the same size for feature splicing or additive fusion.
The grouped interleaved convolution structure is used to replace partial convolutional layers in the deep convolutional network. For example, in the segmentation model shown in Fig. 4, partial convolutional layers are designed as a grouped convolution. In this paper, the 6 convolutional layers (5, 7, 10, 12, 15, and 17) of the network are transformed into grouped convolutional layers, and the other layers are not changed. The specific model structure is shown in Fig. 5.
3D-GFCN takes 3D images as input, after convolution feature extraction, average pooling down-sampling, bi-linear interpolation up-sampling, and FCCRF post-processing optimization, it outputs predicted segmented images of multi-target regions. The specific steps are as follows:
-
(1)
Feature extraction
3D-GFCN uses 3D convolution operation to extract the target features of individual slices and the associated information between adjacent slice features. The feature extraction formula of convolution layer is:
$$\begin{aligned} x_j^{i+1}=\mathrm{Relu}\left( \sum _{m=0}^{M-1}w_{jm}^{i+1}\bigotimes x_m^i+b_j^{i+1}\right) , \end{aligned}$$(2)where \(x_j^{i+1}\) represents the \(j\mathrm{th}\) feature map in \((i+1)\mathrm{th}\) layer, and \(\mathrm{Relu}(\cdot )\) is the activation function. M is the number of feature maps in \(i\mathrm{th}\) layer, and \(w_{jm}^{i+1}\) is the \(m\mathrm{th}\) convolution kernel weight of the \(j\mathrm{th}\) feature map in \((i+1)\mathrm{th}\) layer. \(x_m^i\) is the \(m\mathrm{th}\) feature map in \(i\mathrm{th}\) layer, and \(b_j^{i+1}\) represents the offset of the \(j\mathrm{th}\) feature map in \((i+1)\mathrm{th}\) layer. \(\bigotimes \) represents 3D convolution operation. In practice, the convolution kernel usually use \(3\times 3\times 3\) size and \(1\times 1\times 1\) size, of which \(3\times 3\times 3\) convolution kernel is used for image feature extraction, and \(1\times 1\times 1\) convolution kernel is used to adjust the number of feature map channels of the input and output in convolutional layer, and fuse the feature information between the feature map channels, to facilitate the feature fusion between feature maps and the classification of pixels.
-
(2)
Average pooling down-sampling
After the feature extraction of 3D FCN, the down-sampling adopts the average pooling operation, which increases the receptive field and reduces the model parameters. The down-sampling formula for the feature map is:
$$\begin{aligned} t_{ij}^{x,y,z}=\frac{1}{P\times Q\times R}\left( \sum _{p=0}^{p-1}\sum _{q=0}^{Q-1}\sum _{r=0}^{R-1}t_{(i-1)j}^{(x_0+p)(y_0+q)(z_0+r)}\right) , \end{aligned}$$(3)where \(t_{ij}^{x,y,z}\) represents the feature value at (x, y, z) of \(j\mathrm{th}\) feature map in \(i\mathrm{th}\) layer, and (P, Q, R) is the size of pooling window in \(i\mathrm{th}\) layer. (p, q, r) is the point coordinates of the pooling window, and \(t_{(i-1)j}^{(x_0+p)(y_0+q)(z_0+r)}\) represents the value at \((x_0+p)(y_0+q)(z_0+r)\) of the \(j\mathrm{th}\) feature map in \((i-1)\mathrm{th}\) layer. \((x_0,y_0,z_0)\) represents the left vertex coordinate in the (P, Q, R) neighborhood with (x, y, z) as the center point.
-
(3)
Interpolation up-sampling
The up-sampling of the feature map in 3D FCN uses bilinear interpolation:
$$\begin{aligned} t_{i-1}=\mathrm{conv3D}\left( \mathrm{Interpolation}(t_i)\right) , \end{aligned}$$(4)where \(t_i\) is the feature map in \(i\mathrm{th}\) layer, and \(\mathrm{conv3D}(\cdot )\) represents the 3D convolution operation. \(\mathrm{Interpolation}(\cdot )\) represents the bilinear interpolation operation and \(t_{i-1}\) is the feature map in \((i-1)\mathrm{th}\) layer. From Eq. (4), the up-sampling method adopted is to first use the bilinear interpolation algorithm to up-sample the feature map, and then perform a 3D convolution operation on the up-sampled feature map.
-
(4)
Feature map size cropping and addition fusion
In the fusion of features with different scales between different levels in FCN with short-connection structure, the purpose of the cropping operation is to keep the size of the up-sampling feature map consistent with the size of the additive fusion feature map. The up-sampling cropping formula for feature map is as follows:
$$\begin{aligned} t'_i=\mathrm{crop}\left( t_i,t_j\right) , \end{aligned}$$(5)where \(\mathrm{crop}(t_i,t_j)\) indicates that the size of the feature map \(t_i\) is cropped to the size of \(t_j\).
After the feature map size is unified through the cropping operation, the high-level feature map is up-sampled and combined with the low-level feature map. The detailed information of the low-level network is merged into the high-level feature map, making the segmentation more refined. The specific formula is as follows:
where \(t_i\) is the feature map in \(i\mathrm{th}\) layer, and \(t_j\) is the feature map in \(j\mathrm{th}\) layer. \(average(\cdot )\) represents the addition and fusion operation of feature map, and f is the fused feature map.
-
(5)
Fully connected condition random field post-processing
In practice, fully connected condition random field can be used as a processing tool after segmentation to optimize the segmentation results in the case of poor continuity and unclear segmentation boundary. The structure of FCCRF is shown in Fig. 6.
In Fig. 6, \((x_0,x_1,\ldots ,x_n)\) represents the observation sequence; \((y_0,y_1,\ldots ,y_n)\) represents the classification and labeling sequence, which is a finite state set. \(h(y_k,X)\) is a univariate potential energy function, which is used to construct the relationship between the observation sequence and the classification label; \(g(y_k,y_{k+1},X)\) is a binary potential energy function, which is used to construct the relationship between adjacent classification points in the sequence.
The conditional random field posterior probability model P(X|I) is defined as follows:
where X is the pixel classification set, and I is the original image. E(X|I) is the energy function, and Z(I) is the normalization factor. Conditional random field decision-making is to maximize the posterior probability of the predicted segmentation probability under the input image, and obtain the optimal solution by minimizing the energy function.
The mathematical expression [27] of the energy function:
In Eq. (8), \(\varphi _u(z_i)=-\log P(z_i||I)\) is a univariate potential energy function, \(p(z_i||I)\) is the probability that the pixel i belongs to the category \(z_i\), and \(\varphi _p(z_i,z_j)\) is a binary potential energy function. The definition of \(\varphi _p(z_i,z_j)\) is as follows:
In Eq. (9), \(u(z_i,z_j)\) is a label consistent function. According to the Potts model [28], only when \(z_i\ne z_j\), \(u(z_i,z_j)=k^0\), where \(k^0\) is the consistent matrix, which is a learnable parameter matrix. Otherwise, it is 0, which means that only pixels assigned with different labels are penalized. \(g(f_i,f_j)\) is the Gaussian kernel function [29]:
where P represents position information, and I represents color information. \(\theta _{\alpha }\) is distance similarity parameter, and \(\theta _{\beta }\) is color similarity parameter. \(\theta _{\gamma }\) is smoothness parameter, \(k^1\) is bilateral kernel weight, and \(k^2\) is spatial kernel weight.
According to the definition of Gaussian kernel function, the energy function punishes the pixels assigned different labels according to the spatial distance and color difference, that is, encourages the pixels assigned different labels to have larger spatial distance and color difference, so as to achieve the purpose of optimal segmentation.
Using FCCRF as a post-processing tool for the segmentation model can make the segmentation boundary clearer. The expression is as follows:
where f is the feature map after the fusion of the high-level layer and the low-level layer, and I is the output image. \(\theta \) is the hyperparameter set of FCCRF, \(\mathrm{FCCRF}(\cdot )\) is the FCCRF processing module, and O is the predicted segmentation output.
The post-processing of FCCRF receives the input of the original image and the basic FCN network prediction segmentation image, and optimizes the pixel classification results by calculating the Gaussian penalty function between the pixels assigned with different labels, and finally gets more refined predicted segmentation image.
In the model shown in Fig. 4, the FCCRF is embedded between the last \(1\times 1\) convolution layer and the prediction output layer, as the optimization step of the segmentation results of the basic FCN model.
3 The Learning Algorithm
In this paper, the Dice loss function, which can reduce the influence of background pixels, is used as the model training objective function, and the learning algorithm of the 3D-GFCN model is designed using the Mini-Batch Gradient Descent (MBGD) strategy, and the information processing flow and steps of the training algorithm are established.
3.1 The Design of Loss Function
In complex image segmentation, where the morphological features of the target image are not significant, the quantity imbalance of background pixels and target pixels has a greater impact on the segmentation results. Dice loss function [30] which can ignore the influence of background pixels when used in image segmentation, effectively solve the problem of the imbalance between the background pixels and the target pixels, and make the network have better convergence. In this paper, the Dice loss function is used as the training objective function, which is defined as follows:
In Eq. (12), p is the prediction segmentation, and g is the binary label image. \(\theta _c\) is the training parameter of FCN, and \(\theta _k\) represents the 3 parameter matrices \((k^{(0)},k^{(1)},k^{(2)})\) of FCCRF. \(p\cdot g\) is the matrix dot multiplication operation, and the result represents the predicted segmentation area of the target. \(\mathrm{sum}(\cdot )\) is the summation operation of image gray. The gray level Intersection-over-Union (IoU) between the predicted segmentation image and the label image is used to represent the segmentation accuracy, and the segmentation accuracy is improved by minimizing the segmentation error.
The Dice loss converges quickly in the early stage of training, but when the network training iteration reaches a certain level, because the numerator in Eq. (12) becomes larger, the Dice loss decreases rapidly, which makes it difficult to further improve the training accuracy. To make the network converge to the optimum as soon as possible, in this paper, the binary cross-entropy loss [31] is introduced into the loss function, which is as follows:
In Eq. (13), p is predicted segmentation, and g is label segmentation. \(\theta _c\) represent the training parameters of FCN, and \(\theta _k\) represents label consistent matrix and Gaussian kernel weight of FCCRF. The coefficient \(\beta \) is used to balance image background loss and target loss. The loss function is set as follows:
In Eq. (14), the coefficient \(\alpha \) is used to balance the Dice loss and the binary cross-entropy loss. In the experiment, \(\alpha =e_c/e_t\), where \(e_c\) is the current number of training times, and \(e_t\) is the total number of training times. \(\alpha \) can determine that Dice loss is the main factor in the early stage of network training, and the binary cross-entropy loss is the main factor in the later stage of training.
3.2 Algorithm Implementation
In this paper, the mini-batch gradient descent (MBGD) algorithm with few training iterations and stable convergence is used to train the 3D-GFCN model. The specific algorithm process is as follows:
Step 1: The network training parameters are determined. The 3D-GFCN model training parameters include FCN model parameters and FCCRF parameters. The parameter vector is expressed as follows:
where \(\theta ^{t-1}\) is the network training parameter at time \((t-1)\), \((w_0^{t-1},w_1^{t-1},\ldots ,w_m^{t-1})\) is the set of convolution kernel weight, and \((b_0^{t-1},b_1^{t-1},\ldots ,b_m^{t-1})\) is the bias set. \(k^0,k^1 \mathrm{and} k^2\) are the consistent matrix weight, bilateral kernel weight, and spatial kernel weight of FCCRF, respectively.
Step 2: Randomly sample N 3D image slices \(x_n(n=1,2,\ldots ,N)\), and obtain N corresponding label samples \(g_n\).
Step 3: The predicted segmentation image is obtained:
Step 4: The gradient of the network loss value is calculated:
Step 5: The MBGD algorithm is used to optimize the update of the parameters:
Step 6: If the network converges to the optimal state, the training ends, otherwise it returns to Step 2.
4 Simulation Experiment and Result Analysis
In this section, the experimental data set is composed of 3D CT abdominal medical images. Through the preprocessing of converting the original DCM value into CT value, image normalization and histogram equalization, the proposed method is used to segment the liver and other organs, and the verification experiment and result analysis are carried out. At the same time, the mainstream image segmentation method 2D-FCN, 3D-FCN, 3DFCN-WC and SegNet are selected as the comparative models. Based on the experimental results, the advantages and limitations of different methods are analyzed.
4.1 The Experiment Data Set
In this paper, the medical image public data set 3Dircadb are selected as the experimental data set. The data set contains 20 types of abdominal CT images with segmentation label, including lungs, kidneys, spleen, liver and other organs. 20 groups of individual samples (10 groups of males, 10 groups of females) are extracted from the data set, and the original abdominal images and images with segmentation annotations were selected from each group of samples.
The experimental data set contains a total of 5510 CT image slices. There are 2755 original CT slices and 2755 labeled CT slices, each with a size of \(512\times 512\). The first to sixteenth groups of abdominal slices were used as the training set, the 17 and 18 groups of slices were used as the validation set, and the 19 and 20 groups of abdominal slices were used as the test set. The data set is imbalanced and small-scale, and the specific distribution of the sample is shown in Table 1.
4.2 Data preprocessing
-
(1)
DCM value converted to CT value
The data set stores CT slices in the original DCM format. The DCM pixel value slices are converted into CT values that can reflect the density of different tissues in the abdomen image for image segmentation processing. The conversion formula is as follows:
$$\begin{aligned} \mathrm{Hu}=\mathrm{pixel}\times \mathrm{slope}+\mathrm{intercept}. \end{aligned}$$(19)where Hu represents the CT value, and pixel represents the DCM pixel value. The parameter slope and parameter intercept are obtained from the label information of the DCM image.
-
(2)
Normalization and histogram equalization processing of image
Due to the wide range of image CT value distribution, the boundary of each tissue is not clear. It is necessary to perform windowing processing on the abdominal image, that is, to determine the median and range of the image CT value. If the CT value is less than this range, it is set to 0; If it is in this range, the original value is retained and the CT value is normalized; If it is larger than this range, it is set to 1. In this experiment, the median value is 100 and the range is 500. At the same time, the original CT image is gray and low contrast. Through histogram equalization, the gray level of CT image can be adjusted to enhance image contrast.
4.3 The Model Structure and Parameters are Set
In the experiment, the model structure and detailed parameters are shown in Table 2.
From Table 2, GroupConv represents the grouped convolution module, and the size of the convolution kernel multiplied by 3 represents that the grouped convolution module divides the input feature map into three groups according to the number of channels. Each group of convolution kernels uses \(3\times 3\times 3\) convolution, and the stride is \(1\times 1\times 1\).
4.4 The Experiment Result and Analysis
In the experiment, the MBGD algorithm is used for training. The training epoch is set to 8, the number of iterations is set to 105, and the learning rate of the MBGD optimizer is 0.01. The balance coefficient \(\alpha \) of the binary cross-entropy loss function is set to 0.1, and the hyperparameters of FCCRF are set to \(\theta _{\alpha }=10\),\(\theta _{\beta }=20\),\(\theta _{\gamma }=10\). The experimental environment is in Linux system, and NVIDIA GeForce RTX 2080 Ti GPU.
Pixel accuracy (PA), Class pixel accuracy (CPA), and Intersection over Union (IoU) are used to evaluate the segmentation results. The segmentation evaluation index and training speed of this method based on the experimental results are shown in Table 3.
The segmentation results of the segmentation model on the test set are shown in Fig. 7.
In Fig. 7, there are two sets of 3D data segmentation. Each set consists of 8 slices. The first row in each set are the original abdominal images, and the second row are the gold standard, which is the result of manual segmentation by experts. The third row are the predicted segmentation results of the model in this paper.
It can be seen from Table 3 and Fig. 7 that the liver region segmented by the model in this paper has a small difference in volume and shape from the expert manual segmentation results, and the boundary segmentation of liver organs is smoother, indicating that the model segmentation effect is good.
4.5 The Contrast Experiment and Analysis
To evaluate the performance of 3D-GFCN in segmentation accuracy and training speed, 2D-FCN [32], 3D-FCN [33], 3DFCN-WC [34], SegNet [35] are selected as contrast models. Compared with the proposed model, the 2D-FCN model is different in the dimensions of the convolution kernel. It uses a 2D convolution kernel, the input image is 2D, and other structures are the same as the proposed model. The 3D-FCN model is a fully convolutional network model without grouped convolution structure. 3DFCN-WC is a fully convolutional network model with a grouped convolution structure but no FCCRF as a post-processing. The SegNet is similar to FCN. Its feature extraction and down-sampling part is the first 13 layers of the VGG16 model. Each down-sampling layer corresponds to a deconvolution layer. The size of the convolution kernel is set to \(3\times 3\), and the maximum pooling operation is adopted. The feature map is up-sampled using the up-pooling index method. The training parameter settings of the 4 contrast models, as well as the training set, validation set, and test set are the same as the proposed model.
Pixel accuracy (PA), Class pixel accuracy (CPA), and Intersection over Union (IoU) are used as evaluation indicators for segmentation results. The segmentation evaluation index and training speed of the five models based on the experimental results are shown in Table 4.
From Table 4, compared with the 3D-FCN model, the training speed of proposed model increased by 12.12%, but the segmentation index of the proposed model is slightly lower. The reason is that the proposed model used grouped convolution, which leads to partial loss of information between the channels of feature map, but the training speed is improved. Compared with the 3DFCN-WC model, the proposed model used FCCRF as a post-processing tool, so the segmentation accuracy has been greatly improved, and the four segmentation accuracy indexes were improved by more than 2.1%. Comparing the SegNet and the 2D-FCN, for the four evaluation indexes, the proposed model has greatly advantages, but the training speed of the proposed model is lower. The reason is that the 2D segmentation model needs fewer parameters to calculate, the training time is short, and the convergence speed is fast. However, in the four evaluation indicators, the proposed model has greatly advantages.
For the ability to distinguish between background pixels and target pixels, the confusion matrix of the proposed model is shown in Table 5, and the experimental segmentation index are shown in Table 6.
From Table 5, since background pixels account for most of the image, the recognition rate of background pixels is high, and some liver pixels are mistakenly classified as background pixels. From Table 6, 91.35% of all liver pixels are correctly identified, indicating that the model segmentation effect is good. 95.77% of all the pixels recognized as liver are real liver pixels, indicating that the model has a strong ability to recognize liver pixels. The IoU index of liver pixels is 87.82%. The main factor affecting it is that the number of liver pixels is relatively small compared to the entire image, and a small number of pixels are mistakenly classified as background pixels, but this is still a good result.
Comprehensive analysis shows that the proposed method has overall advantages over the other four comparison models, and has good applicability to the fine segmentation of 3D abdominal CT images with insignificant morphological features, overlapping boundaries and local occlusion.
5 Conclusion
In this paper, aiming at the utilization of the correlation information between adjacent slices and the optimization of segmentation results in 3D image segmentation, a 3D grouped full convolutional network model using fully connected conditional random field as a post-processing tool is proposed. In mechanism, 3D-GFCN can use the ability of 3D convolution expressing related information of adjacent slices, contextual information association and probability exploration mechanism of FCCRF, and shortcut-connection structure of FCN, which can effectively improve the accuracy of multi-target instance segmentation of complex 3D image. At the same time, the grouped convolution structure is adopted, which can greatly reduce the model parameters and reduce the complexity of the 3D convolution operation. The 3D abdominal CT images segmentation experiments based on the 3Dircadb data set show that the proposed method has a good performance in segmentation accuracy and training speed. The CPA reached 95.3%, IOU 87.8% and accuracy 91.3%, all of which achieved good segmentation accuracy. At the same time, the training speed is increased by 12.12% compared to 3D-FCN. It shows 3D-GFCN has good applicability to solve the image segmentation problem with insignificant morphological features and weak spatial information correlation of the target image, and improves the limitations and shortcomings of the comparison method in solving the above problems. It can provide a novel and effective deep learning model for segmenting 3D images. However, the information processing mechanism and algorithm process of this method are more complicated, and the image semantic knowledge and structural features are less used. From the experimental results, the number of image background pixels accounts for too much compared with the pixels in the target area, and the imbalanced quantity of samples in the data set still have certain impact on the segmentation accuracy. How to optimize the model and algorithm, improve the distinguishing ability of the target image and background image features, and embed the scene semantic feature knowledge into the segmentation model, will be the important work in the next stage.
Availability of data and materials
Not applicable.
Abbreviations
- 3D-GFCN:
-
3D grouped fully convolutional network
- FCN:
-
Fully convolutional network
- FCCRF:
-
Fully connected conditional random field
- CT:
-
Computer tomography
- MRI:
-
Magnetic resonance imaging
- DCNN:
-
Deep convolutional neural network
- 3D CNN:
-
3D convolutional neural network
- MBGD:
-
Mini-batch gradient descent
References
Minaee, S., Boykov, Y.Y., Porikli, F., Plaza, A.J., Kehtarnavaz, N., Terzopoulos, D.: Image segmentation using deep learning: a survey. IEEE Trans. Pattern Anal. Mach. Intell
Chen, C., Qin, C., Qiu, H., Tarroni, G., Duan, J., Bai, W., Rueckert, D.: Deep learning for cardiac image segmentation: a review. Front. Cardiovasc. Med. 7, 25 (2020)
Zhou, Z., Siddiquee, M.M.R., Tajbakhsh, N., Liang, J.: Unet++: A nested u-net architecture for medical image segmentation, In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer, pp. 3–11, (2018)
Gu, Z., Cheng, J., Fu, H., Zhou, K., Hao, H., Zhao, Y., Zhang, T., Gao, S., Liu, J.: Ce-net: context encoder network for 2d medical image segmentation. IEEE Trans. Med. Imaging 38(10), 2281–2292 (2019)
Milletari, F., Navab, N., Ahmadi, S.-A.: V-net: Fully convolutional neural networks for volumetric medical image segmentation, In: 2016 Fourth International Conference on 3D Vision (3DV), IEEE, pp. 565–571 (2016)
Chen, J., Yang, L., Zhang, Y., Alber, M., Chen, D.Z.: Combining fully convolutional and recurrent neural networks for 3d biomedical image segmentation, In: Advances in Neural Information Processing Systems, pp. 3036–3044 (2016)
Baumgartner, C.F., Koch, L.M., Pollefeys, M., Konukoglu, E.: An exploration of 2d and 3d deep learning techniques for cardiac mr image segmentation, In: International Workshop on Statistical Atlases and Computational Models of the Heart, Springer, pp. 111–119, (2017)
Zhang, Y., Liu, Y., Cheng, H., Li, Z., Liu, C.: Fully multi-target segmentation for breast ultrasound image based on fully convolutional network. Med. Biol. Eng. Comput. 58(9), 2049–2061 (2020)
Tian, Y., Dehghan, A., Shah, M.: On detection, data association and segmentation for multi-target tracking. IEEE Trans. Pattern Anal. Mach. Intell. 41(9), 2146–2160 (2018)
Dinghan, W., Guilan, F., Xiong, W., Yufeng, W., Maohua, D.: Research on image segmentation algorithm based on features of venous gray value. Opto-Electron. Eng. 45(12), 180066–1 (2018)
Makandar, A., Halalli, B.: Threshold based segmentation technique for mass detection in mammography. J. Comput. 11(6), 472–478 (2016)
Gupta, D., Anand, R.: A hybrid edge-based segmentation approach for ultrasound medical images. Biomed. Signal Process. Control 31, 116–126 (2017)
Kashyap, R., Gautam, P.: Modified region based segmentation of medical images. In: 2015 International Conference on Communication Networks (ICCN), IEEE, pp. 209–216 (2015)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. Adv. Neural. Inf. Process. Syst. 25, 1097–1105 (2012)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition, In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Ren, S., He, K., Girshick, R., Sun, J.: Faster r-cnn: towards real-time object detection with region proposal networks. Adv. Neural. Inf. Process. Syst. 28, 91–99 (2015)
Hesamian, M.H., Jia, W., He, X., Kennedy, P.: Deep learning techniques for medical image segmentation: achievements and challenges. J. Digit. Imaging 32(4), 582–596 (2019)
Despotović, I., Goossens, B., Philips, W.: Mri segmentation of the human brain: challenges, methods, and applications. Comput. Math. Methods Med., (2015)
Kamnitsas, K., Chen, L., Ledig, C., Rueckert, D., Glocker, B.: Multi-scale 3d convolutional neural networks for lesion segmentation in brain mri. Ischemic Stroke Lesion Segment. 13, 46 (2015)
Mlynarski, P., Delingette, H., Criminisi, A., Ayache, N.: 3d convolutional neural networks for tumor segmentation using long-range 2d context. Comput. Med. Imaging Graph. 73, 60–72 (2019)
Valverde, S., Cabezas, M., Roura, E., González-Villà, S., Pareto, D., Vilanova, J.C., Ramió-Torrentà, L., Rovira, À., Oliver, A., Lladó, X.: Improving automated multiple sclerosis lesion segmentation with a cascaded 3d convolutional neural network approach. Neuroimage 155, 159–168 (2017)
Wang, L., Xie, C., Zeng, N.: Rp-net: a 3d convolutional neural network for brain segmentation from magnetic resonance imaging. IEEE Access 7, 39670–39679 (2019)
Dhia, H., Ghani, R.F.: A proposed method for scale drawing calculating depending on line detector and length detector. Iraqi J. Comput. Sci. Math. 2(2), 6–17 (2021)
Lafferty, J., McCallum, A., Pereira, F.C.: Conditional random fields: Probabilistic models for segmenting and labeling sequence data
Soler, L., Hostettler, A., Agnus, V., Charnoz, A., Fasquel, J., Moreau, J, Osswald, A., Bouhadjar, M., Marescaux, J.: 3d image reconstruction for comparison of algorithm database: a patient specific anatomical and medical image database, IRCAD, Strasbourg, France, Tech. Rep.
Kim, D.-S., Kim, Y.-H., Park, K.-R.: Semantic segmentation by multi-scale feature extraction based on grouped dilated convolution module. Mathematics 9(9), 947 (2021)
Rao, C.H., Naganjaneyulu, P., Prasad, K.S.: Brain tumor detection and segmentation using conditional random field, In: 2017 IEEE 7th International Advance Computing Conference (IACC), IEEE, pp. 807–810 (2017)
Gridchyn, I., Kolmogorov, V.: Potts model, parametric maxflow and k-submodular functions, In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2320–2327 (2013)
Murua, A., Stanberry, L., Stuetzle, W.: On Potts model clustering, kernel k-means and density estimation. J. Comput. Graph. Stat. 17(3), 629–658 (2008)
Huang, Q., Sun, J., Ding, H., Wang, X., Wang, G.: Robust liver vessel extraction using 3d u-net with variant dice loss function. Comput. Biol. Med. 101, 153–162 (2018)
Ruby, U., Yendapalli, V.: Binary cross entropy with deep learning technique for image classification. Int. J. Adv. Trends Comput. Sci. Eng. 9(10)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation, In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Fechter, T., Adebahr, S., Baltas, D., Ben Ayed, I., Desrosiers, C., Dolz, J.: Esophagus segmentation in CT via 3d fully convolutional neural network and random walk. Med. Phys. 44(12), 6341–6352 (2017)
Wang, B., Lei, Y., Tian, S., Wang, T., Liu, Y., Patel, P., Jani, A.B., Mao, H., Curran, W.J., Liu, T., et al.: Deeply supervised 3d fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation. Med. Phys. 46(4), 1707–1718 (2019)
Badrinarayanan, V., Kendall, A., Cipolla, R.: Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017)
Acknowledgements
The authors would like to thank to the anonymous reviewers and editor for their insightful comments.
Open Access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Funding
This work was supported by the Shandong University of Science and Technology Research Fund under Grant 2019TDJH102.
Author information
Authors and Affiliations
Contributions
JY designed the study and analyzed the data. ZZ contributed to the study design and took the lead in the manuscript writing. SX supervised the study design and helped in the writing of the final draft of the manuscript. RY and KL made critical revisions of the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have reviewed the final version of the manuscript and approve it for publication.
Conflict of interest
The authors declared that they have no conflicts of interest to this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yin, J., Zhou, Z., Xu, S. et al. A 3D Grouped Convolutional Network Fused with Conditional Random Field and Its Application in Image Multi-target Fine Segmentation. Int J Comput Intell Syst 15, 11 (2022). https://doi.org/10.1007/s44196-022-00065-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44196-022-00065-w