Abstract
Arthropods serve as efficient carriers for various life-threatening infections parasites, viruses and other harmful organisms throughout the world. In this study, Al-Aqiq Governorate of Al-Baha city served as the site where tick burden on camels (Camelus dromedarius) was assessed. The aim of the present investigation was to determine the diversity and prevalence of ticks. A total of 800 ticks, 756 adults and 44 nymphs were gathered and identified. Tick DNA was obtained from camels by utilizing commercially available kits. Polymerase chain reaction (PCR) was done on recovered DNA utilizing 12S rDNA and 16S ribosomal DNA fragments to analyze and identify the ticks. Three species of hard ticks were found in the two genera Hyalomma and Rhipicephalus. Hyalomma dromedarii (81.81%) and H. rufipes (17.35%) were the two species with the highest prevalence found on the domestic animal hosts, whereas R. sanguineus (had the lowest prevalence of 0.82%). This is a preliminary report of DNA barcode of tick species of camels (Camelus dormedarius) from Al-Baha in KSA and the Arabian Peninsula, which is a significant step towards broader phylogenetic studies on tick samples from this region. The results of our study provide valuable insights for healthcare administrators and government authorities regarding the frequency of hard ticks on mammalian hosts in Al-Aqiq Governorate of Al-Baha City, Saudi Arabia. This can also assist in regulating tick diseases, in particular during the epidemic time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The Arabian camel and the dromedaries have one hump. Camelus dromedarius is highly valued in the Kingdom of Saudi Arabia (KSA) due to its economic, veterinary, and cultural importance, making it one of the most popular livestock species in the country. Camels are typically raised in hot, dry temperatures in their native desert habitats, which exposes them to significant stress and renders them susceptible to several diseases, mostly due to infestations by external parasites such as hard ticks [1]. Ticks are regarded as significant arthropods that mostly consume blood, particularly due to their function as carriers of diseases. Ticks transmit a wide range of pathogens that are recognized for causing several animal and human diseases globally, ranking second only to mosquitoes in this aspect [2, 3]. Ticks are significant parasites in terms of both economic and public health implications due to their capacity to diminish farm animal productivity and transmit zoonotic diseases. Although ticks are regarded as the primary vector of illnesses damaging cattle globally, ticks and mosquitoes are the most frequent carriers of pathogens that cause human and animal diseases [4]. Ticks are globally acknowledged arthropod vectors of infection-producing pathogens in medicine and animal sciences, presenting a zoonotic and imminent risk to human health [5]. Besides, ticks have secondary effects on their hosts during feeding, including the development of anaemia, paralysis, immunosuppression, and the infiltration of secondary bacterial infections at tick bite spots. Therefore, as a result of harmful impacts both directly and indirectly, ticks and the diseases they carry pose a significant risk to the well-being and productivity of domestic animals, as well as the security of our food supply. It has been observed that over 20 species of ticks from the ixodid family have been identified as infesting camels [6, 7]. Tick-borne infections have been increasingly prevalent in human beings as well as animals in Saudi Arabia. This encompasses the spread of several diseases transmitted by ticks, such as Crimean-Congo hemorrhagic fever, a zoonotic disease [8,9,10], Alkhurma virus, which causes a tick-borne viral disease known as Flavivirus [11], and theileriosis and babesiosis in sheep, horses, and camels [12,13,14,15,16]. In the Arabian Peninsula, camel species are estimated to be 1.6 million, with 53% of them located in Saudi Arabia [17]. Hard ticks, belonging to the family Ixodidae, are found all over the world and tend to prefer temperate zones more than soft ticks, which belong to the family Argasidae. Hyalomma ticks exhibit a wide geographical range, encompassing the Middle East, the Arabian Peninsula, and multiple Asian nations. The tick species frequently encountered are H. dromedarii, H. schulzei, H. anatolicum, Boophilus kohlsi, Rhipicephalus turanicus, and H. impeltatum in the kingdom [1]. Because of their remarkable adaptability to human and animal settings, ticks have assumed a prominent role in public health and economic concerns [18]. Therefore, the key component of this investigation was to identify morphological and molecular characteristics of tick species that infest Dromedary camels (Camelus dromedarius) in the Al-Aqiq province of Al-Baha City, located in the southwestern region of Saudi Arabia. As far as we are aware, it is the primary document of DNA barcode of H. dromedarii ticks from the Kingdom and Arab region. This represents a major milestone in the pursuit of more extensive phylogenetic investigations including tick samples from this area. These findings assist cattle as well as people wellbeing services, and official authorities, in acquiring a comprehensive understanding of the infestation of mammalian hosts by hard ticks. This can also help to prevent outbreaks of tick diseases.
2 Methods and materials
2.1 Ethical approval
The ethical Committee of the Department of Biology, Faculty of Science, Al-Baha University examined and approved this study, following the guidelines set by the Deanship of Scientific Research, Al-Baha University. The local government was permitted to conduct this study at the designated study sites, with authorization obtained from the District Veterinary Officers et al.—Baha University and the Al-Aqiq District Councils. Verbal consent was acquired from the animal owners before gathering data. This research has been done with the explicit consent of owners, district councils of investigation locations, Department of Biology et al.—Baha University’s Faculty of Science.
2.2 Study sites and sample collection
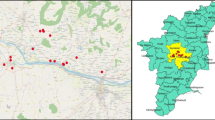
The objective of this study was to investigate the diversity and prevalence of ticks in Al-Baha region. The sample of ticks was collected from the animal carcasses from several camel breeding yards. A total of 121 camels (755 adults and 44 nymphal) were collected between June 2021 and July 2022, representing diverse maturation phases. The camels were first domesticated in Al-Aqiq Governorate, which is situated south-western region (Fig. 1). Latitudes 20° 16′ 6.6′′ N and 41° 38′ 57.48′′ E, where Al-Baha University and King Saud Bin Abdulaziz Airport are situated. The general climate belongs to the arid zone category. The minimum temperature is 12 °C and the maximum temperature is 23 °C. The relative humidity goes from 52 to 67% [19]. Ticks are extremely vulnerable to climate change due to the length of their life-cycle stages when they are not attached to a host. During these stages, processes like development and survival are greatly influenced by temperature and moisture levels [20]. The abundance of a species is likely to fluctuate depending on how temperature affects its development and mortality at different phases of its life cycle, particularly when considering seasonal changes [21,22,23]. These three basins are located in Al-Baha province, which is the smallest province in the Saudi Arabian monarchy [24]. Despite being substantially higher than the Saudi average, rainfall in Al-Baha province averages between 200 and 600 mm per year [25, 26]. Ticks were taken off from two body areas (breast, and abdomen), while the camels were being fed. Ticks were removed from animals using forceps to grasp them. The tick attaches securely to its scutum and mouthparts, close to the skin and then placed in tubes that had been marked and filled with 70% ethanol. All of the camels were of different genders. Once the species of the ticks had been determined, the samples were retained in 70% molecular grade ethanol and stored at −20 °C for further molecular characterization.
A map of Saudi Arabia displaying the Al-Aqiq governorate (blue circle), which is located in Al Baha, in the country’s southwest and from where tick samples were collected from the camel farms [27]
2.3 Morphological identification of ticks
All ticks were identified morphologically using a stereo microscope as per their morphological features (based on: the presence or absence of festoons, eye existence or absence, mouthpart size, scutum, Adanal plate shape ends, Spiracle regions, Subanal plate, etc..) with the aid of keys and guides as reported previously [6, 28].
2.4 DNA extraction and molecular identification
Prior to extracting tick DNA, every tick underwent a five-minute cleaning process that included spinning at 13,000 × rpm in 1.5 ml of freshly made phosphate-buffered saline (PBS). The clean ticks were crushed Individually with autoclaved mortar and pestle to get homogenate. DNA has been extracted from 40 ticks, (5–8 ticks from each species) using the mini-prep DNA isolation kit (GeneAll® Exgene™ Clinic SV DNA Isolation Kit, Seoul, South Korea), with the suitable modifications: tick samples were digested in 200 µl of lysis buffer(CL), 20 µl of proteinase K for 16 h at 56 °C on a heat-block. The subsequent steps were carried out according to manufacturer’s guidelines and the whole DNA was extracted and collected in 70 µl of elution solution (Tris buffer with a pH of 8.5, heated to a temperature of 70 °C). For subsequent PCR and sequence analysis, we used gDNA from three species: Hyalomma dromedarii, R. sanguineus, and Hyalomma rufipes. The tick species were verified with the use of conventional PCR targeting 16S rRNA and 12S rDNA as DNA barcoding genes to identify specific tick species.
For tick identification specific primers amplifying 12S ribosomal DNA (12S rDNA) gene, and a 16S ribosomal DNA (16S rDNA) gene [30, 31], using the primers listed in Table 1. Amplification of these genes was achieved using BioMix Red, 2x (Bioline, BIO-25006, Meridian Bioscience, Germany). The PCR amplifications were performed in a total reaction volume of 30 µl with the following mixture: 17.5 µl nuclease-free H2O, 5 µl of DNA template, 1 µl of each primer, and 15.5 µl BioMix Red. The reaction was done on thermal cycler (Cat No./ID: BIO-25006, Veriti 96-Well Thermal Cycler, Applied Biosystems, Waltham, MA) under these conditions: a denaturation step at 95°C for 1 min, followed by 35 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 10 s, with 72 °C for 5 min as the final step. A UV transilluminator was used to observe the PCR products that were produced on 1.5% agarose gels in 1 × TBE buffer (SERVA Electrophoresis, Heidelberg, Germany) dye with gel red (Phonex Research Products, Candler, USA). There was PCR-grade water used as a negative control in each reaction and a positive control in everyone. Macrogen Ltd. of Seoul, South Korea, bidirectionally sequenced the PCR material. To find sequence similarities, acquired DNA sequences were matched with sequences in the GenBank database using the Basic Local Alignment Search Tool (BLAST). (https://blast.ncbi.nlm.nih.gov (accessed on 23 December 2022). Using the software Sequence Scanner Version 2.0, the sequencing chromatogram's quality was assessed (Applied Biosystems, Foster City, CA). Using Bioedit version 7.2.5, a consensus nucleotide sequence was created by aligning the reverse complement and forward nucleotide sequences separated by the reverse and forward primer sequences (Ibis Biosciences, Carlsbad, CA). BLASTn was employed to investigate nucleotide identity by comparing the consensus nucleotide sequence with other nucleotide sequences in the GenBank database.
2.5 Data analysis
The information was entered into a Microsoft Excel spreadsheet and properly coded. SPSS version 16 was utilized for the data analysis. Descriptive statistics were employed in this data analysis to establish the prevalence of camel tick infection. Distribution and risk factors, such as sex, were tested using chi-square. 95% confidence intervals and P 0.05 were used in every instance to determine significance. One-way ANOVA has been carried out to analyze mean tick burden among variables (e.g., Tick species stage, tick sex and sum of squares, degrees of freedom, mean square, and tick control frequency). To obtain statistical significance a criterion was set at a p-value below 0.05. The sequences obtained from this study were searched for similar sequences in the GenBank1 database using BLASTn (www.ncbi.nlm.nih.gov/BLAST). The Clustal W program was utilized to align the sequences. Nucleotide alignments were conducted based on these alignments, and Phylogenetic analyses were carried out using MEGA version 7.0, which may be found at https://www.megasoftware.net/. The phylogenetic tree was constructed using the Neighbor-Joining method, which relies on the 16S rRNA gene sequences of ticks. The bootstrap test quantifies the fraction of duplicate trees related to taxa that are clustered, and this data is presented alongside the branches.
3 Result
3.1 Ticks collection
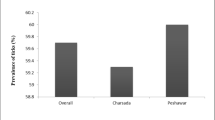
In the present study, 800 ticks out of 121 camels (Camelus dormedarius) (6.61%) were identified to be infected with ixodid ticks. Arabian camels yielded a collection of 800 tick specimens, consisting of 272 females, 484 males, and 44 nymphs. Ticks, as a whole, were determined to be part of the Ixodidae family. The tick specimens were classified into three species, comprising two genera of Hyalomma and Rhipicephalus. The research area has a recorded total tick prevalence of 98.98% (n = 121). The tick species with the highest prevalence was Hyalomma dromedarii, with a prevalence rate of 81.81%. This was followed by H. rufipes, which had a prevalence rate of 17.35%. The tick species with the lowest prevalence was R. sanguineus, with a prevalence rate of 0.82% (Table 2, Fig. 2). The current study identified Hyalomma dromedarii as the most prevalent and abundant tick species among those collected from Al-Aqiq governorate, accounting for 83.5% of the total. The proportion of each tick species identified is shown in Table 3. H. dormedarii female (n = 67 (85%) was the most prevalent tick species, followed by H. dormedarii-1(n = 34 (79%), while H. rufipes male was the least prevalent with a (n = 8, 18%). There was a significant difference (p < 0.046*) in the prevalence of the different tick species stages collected in camels (Table 4).
A taxonomically and molecularly confirmed Hyalomma dromedarii stereomicroscope was used to photograph the item, as demonstrated in Figs. 2, 3.
Characters of taxonomic importance of ticks species. A Hyalomma dromedarii Adult male. Note the shape of the adanal plates has round ends (yellow arrows) and the Subanal plates alignment is outside the adanal plates (blue arrows); B Rhipicephalus sanguineus. shows Palp articles are all small (blue arrows) and Basis capituli has distinctly angular lateral margins (yellow arrows) C Hyalomma rufipes Adanal plates shape has square ends (yellow arrows) and Subanal plate alignment is with the adanal plates. Subanal plates are distinct (blue arrows); D Hyalomma rufipes display Spiracle areas that have sparse setae
3.2 Molecular identification of ticks
For molecular analysis, a sample of 40 ticks was randomly chosen from each species, with sizes varying from 1 to 7. All 15 samples with questionable CO1 findings had a successful amplification of the 16S rRNA gene. The amplification of a sequence of 16S rRNA, approximately 276 base pairs in length, resulted in the production of the anticipated amplification products (Fig. 4). Moreover, there was 99–100% similarity between the three sequences of three species of ticks sequences in the GenBank.
Displays a 1% agarose gel electrophoresis image that demonstrates the polymerase chain reaction product obtained by amplifying the 16S rRNA gene from ticks' DNA. The anticipated size of the amplicon was 276 base pairs. The 100 base pair ladders are represented by M. Samples 1 to 3 were obtained from H. dromedarii, while samples 4, 5, and 6 were obtained from H. rufipes. Samples 8, 10, and 11 were obtained from Rhipicephalus sanguineus. Sample 12 served as a negative control, lacking DNA
Considering 16S rRNA gene sequences, Hyalomma dromedarii from our research (Gene Bank: OQ923646, OQ923647, OQ923648, OQ923649, OQ923650) showed the closest similarity to Hyalomma dromedarii isolate sequence from Riyadh, Saudi Arabia (GeneBank: MG972372). Hyalomma rufipes from this study (Gene Bank: OQ152526) had the closest similarity to Hyalomma rufipes sequence from South Africa (GeneBank: KU130462) (Fig. 5).
Displays a phylogenetic tree created using the Neighbor-Joining method, which is based on the genetic sequences of the 16S rRNA gene in ticks. The bootstrap test displays the percentage of replicate trees where the related taxa formed clusters adjacent to the branches. The samples sequenced in this study are represented by blue triangles and a red circle
The results of the 12S rRNA gene-amplified PCR product showed that the DNA amplicon size for the reaction was 336 bp, suggesting the positive samples for the target gene (Fig. 6). The nucleotide sequences of ticks obtained in this study were submitted to the GenBank and assigned unique accession numbersOQ161687-OQ161692 and OQ822820-OQ822821, respectively. Additionally, accession number OQ161643 was also provided. Based on the 12S rRNA gene sequences, Hyalomma dromedarii from this study (Gene Bank: OQ822827) showed the closest similarity to Hyalomma dromedarii isolate sequence from Atlanta-USA (GeneBank: AF150036.1). H.marginatum from this study Gene Bank: OQ822820, and OQ822821) showed the closest similarity to Hyalomma rufipes- isolate sequence from Atlanta, Georgi, (GeneBank: AF150033.1), whereas, R. sanguineus sequence from this study were assigned to the (GenBank OQ161643) showed closest similarity to Rhipicephalus sanguineus isolate sequence from U.S, and Portugal, (GeneBank: AF150020.1, MF425943.1) respectively (Fig. 7).
Displays a 1% agarose gel electrophoresis image that demonstrates the polymerase chain reaction product obtained by amplifying the 12S rRNA gene from ticks' DNA. The anticipated size of the amplicon was 336 base pairs. The 100 base pair ladders were labeled as M. Samples 1 to 3 were from H. dromedarii, samples 4, 5, and 6 were from H. rufipes, and samples 7, 8, and 9 were from Rhipicephalus sanguineus. Sample 10 served as a negative control, lacking DNA
Displays a phylogenetic tree created using the Neighbor-Joining method. The tree is based on the genetic sequences of the 12S rRNA gene found in ticks. The bootstrap test displays the percentage of duplicate trees where the connected taxa clustered together, indicated next to the branches. The samples sequenced in this study are represented by green circles, red triangles, and blue squares
Phylogenetic analysis was employed to ascertain genetic correlation among nucleotide sequences acquired herein and reference sequences from GenBank. This analysis was conducted using 16S and 12S rRNA nucleotide sequences of the tick species that were identified. The nucleotide sequences for the 16S gene from each species of tick used in this study were aligned, and the results revealed the complete identity of the sequences. Similar findings were made with the nucleotide sequences of the 12S gene, which were collected from each species of tick. Therefore, to conduct phylogenetic analysis, only one sequence from each tick species was utilized.
4 Discussion
Ticks are second to mosquitoes among rickettsial vectors: bacteria, viruses, and protozoa. The current study involved the morphological identification of ticks that infect camels in Al-Baha. The outcome of this research indicated that three species of hard ticks from two genera infest the camel population, which is similar to earlier investigations in Saudi Arabia [33,34,35]. The identification process relied on morphological characteristics and was conducted using the keys provided in the referenced sources [6]. Further, the identification was authenticated using a DNA molecular technique that involved performing PCR and sequencing of cytochrome oxidase subunit I region of ticks’ mitochondrial DNA, especially targeting the 12S rDNA and 16S rDNA [31]. Several studies have been conducted on tick species of camels from various countries of the world but still very little research work has been conducted in Saudi Arabia. Hence, the research aimed to perform morphological and genetic identification of parasites and determine their prevalence in camels residing in the Baha region. In the current study, we investigated the morphological and molecular characteristics of tick species from camels (Camelus dormedarius). Taxonomic keys were used to define the morphology of the flies, whereas molecular characteristics were identified based on the cytochrome c oxidase subunit-1 region of the mitochondrial DNA of ticks, (12S rDNA) and 16S ribosomal DNA (16S rDNA).
In the present study, the frequency of tick species infection was found to be (81.81%) in Hyalomma dromedarii followed by H. rufipes (17.35%), while, R. sanguineus had the lowest prevalence (0.82%). Ticks which infect camel were Hyalomma dromedarii; R. sanguineus, and Hyalomma rufipes based on morphological features [6, 28]. Ticks from genus Hyalomma are predominant and were established to be a varied group and were reported from all areas, this clearly demonstrates that H. dromedarii adapts well to dry environments [33, 36,37,38]. Our data is in agreement with the reported findings [39,40,41]. The influence of climate change on transmission is anticipated to be particularly notable for vectors when they occur within the boundaries of extreme temperature ranges [42]. Hence, the varied climatic conditions in Saudi Arabia create a favourable habitat for the rapid spread of different tick species. According to Abdullah et al. [43] was found (39.9% and 70.60%) respectively of H. dromedarii. The prevalence of infection was much greater in the current study compared to the findings reported by Abdullah et al. [43]. H. dromedarii infects camels and can also infest other cattle. The same results on tick prevalence have been reported elsewhere [34, 35, 44]. Furthermore, dogs and wild rodents can occasionally serve as hosts for a diverse range of other species [21, 45]. It was discovered that temperature, relative humidity, and rainfall all had an impact on tick dispersal. We herein have concentrated on the species Hyalomma dromedarii because it is readily available for extensive research studies [21]. H. dromedarii can spread pathogens such as Theileria annulata and identified lesser group Rickettsia, which are responsible for mortality in camel species [47]. They recognized various tick-host associations among the 37 tick species and subspecies that infest livestock and wild animals [6, 34].
A recent research study has substantiated that ticks are most abundant throughout the summer season, with the principal period of tick activity coinciding with this time. These findings align with prior studies on the subject [46, 48, 49]. Hyalomma rufipes and R. sanguineus, were also found in both the animal species studied to have (17.35%) and (0.82%) respectively. The ticks were commonly seen infesting camels, water buffaloes, cattle, goats, sheep, donkeys, and horses [44, 50, 51]. From the survey of review of the literature, it was discovered that Amblyomma, Haemaphysalis, Hyalomma, and Rhipicephalus are the four principal genera, each of which contains seventeen species, that infest animals in Saudi Arabia.
In the current study, the first documented occurrence of Hyalomma rufipes in camels (C. dromedarius) was recorded. Al- Baha Province17.35%. However, in contrast, a study in Tabuk in northwestern Saudi Arabia was not in agreement with these results and found that 1% of Hyalomma rufipes (only male) [52]. In addition, Previous research revealed that Hyalomma rufipes are mainly found in Africa and the Arabian Peninsula [53, 54]. Furthermore, literature analysis uncovered that Amblyomma, Haemaphysalis, Hyalomma, and Rhipicephalus are the primary genera consisting of seventeen species that infest the animals in Saudi Arabia. Based on molecular characterization, [43] described two tick species of H. dromedarii and H. excavatum, using COI and 16S analysis. It was suggested that H. dromedarii ticks and camels could potentially be the origins of Q-fever infection in humans and other animals [55]. showed that COI evidence supports the genetic similarity of H. dromedarii ticks in the United Arab Emirates (UAE). However much more investigation is required to comprehend their intricate population structure and to clear up the misunderstandings in the systematic relationships between many of the species that have been previously investigated [56]. According to the findings of this study which are in line with earlier research from Saudi Arabia, three hard tick species from two genera infest camel [57,58,59,60].
To the best of our knowledge, our investigation is the first report on the presence and dispersion of ticks in Al-Baha, Saudi Arabia. High annual rainfall characterizes this semi-arid city. Hyalomma dromedarii made up 95% of the species. Overall, this research establishes the basic knowledge of ticks, which could improve the effectiveness and safety of control programs in arid regions around the world. Conclusively, future research should cover as many different regions as possible of the Al-Baha. This will be a crucial step to bring to the effectiveness of initiatives to manage acaricide resistance and illness.
5 Conclusion
These results demonstrate that H. dromedarii is the predominant ixodid tick from Al-Aqiq Governorate; also, tick species diversity was found to be less in that area. Since H. dromedarii is an important tick vector of pathogens to both humans and animals in Saudi Arabia, further studies on the prevalence of pathogens transmitted by I. ricinus should be done in this area. Nevertheless, this study for the first time confirmed the presence of the tick species. Hence, it is crucial to examine the epidemiological and genetic characteristics of different tick species in other areas of Al-Baha. Our work will provide valuable insights for investigating and developing control measures for tick control mechanisms.
Data availability
The data underlying this article are available in article. The data generated or analyzed during current study are available from corresponding author on request.
References
Al-Shammery KA, Fetoh BEA, Alshammari AM (2011) Differentiation between common tick species using molecular biology techniques in Saudi Arabia. J World Academy Sci Eng Tech 73:305–307
Sonenshine DE, Roe RM (eds) (2013) Biology of ticks, vol 2. Oxford University Press, Oxford
Estrada-Peña A, Venzal JM, Kocan KM, Sonenshine DE (2008) Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci 13(18):6938–6946
Zeman P, Benes C (2013) Spatial distribution of a population at risk: an important factor for understanding the recent rise in tick-borne diseases (Lyme borreliosis and tick-borne encephalitis in the Czech Republic). Ticks Tick-Borne Dis 4(6):522–530
Dantas-Torres F, Chomel BB, Otranto D (2012) Ticks and tick-borne diseases: a one health perspective. Trends Parasitol 28(10):437–446
Hoogstraal H, Wassef HY, Büttiker W (1981) Ticks (Acarina) of Saudi Arabia. Fam. Argasidae Ixodidae. Fauna of Saudi Arabia 3:25–110
Silaghi C, Beck R, Oteo JA, Pfeffer M, Sprong H (2016) Neoehrlichiosis: an emerging tick-borne zoonosis caused by Candidatus Neoehrlichia mikurensis. Exp Appl Acarol 68(3):279–297
El-Azazy O, Scrimgeour E (1997) Crimean-Congo haemorrhagic fever virus infection in the western province of Saudi Arabia. Trans R Soc Trop Med Hyg 3:275–278
Perveen N, Kundu B, Sudalaimuthuasari N, Al-Maskari RS, Al-Deeb MSB, MA, (2023) Virome diversity of Hyalomma dromedarii ticks collected from camels in the United Arab Emirates. Vet World 16(3):439–448
Abdulall AK, Zaki AM, Raslan F, Bendary HA (2023) Prevalence of tick- borne viruses from ticks breeding on camels imported to Egypt during the period from January 2019 to April 2021. AIJPMS 3(1):64–77
Charrel RN, Zaki AM, Attoui H, Fakeeh M, Billoir F, Yousef AI, de Chesse R, De Micco P, Gould EA, de Lamballerie X (2001) Complete coding sequence of the Alkhurma virus, a tick-borne flavivirus causing severe hemorrhagic fever in humans in Saudi Arabia. BBRC 287(2):455–461
El-Azazy O, El-Metenawy T, Wassef H (2001) Hyalomma impeltatum (Acari: Ixodidae) as a potential vector of malignant theileriosis in sheep in Saudi Arabia. Vet Parasitol 99(4):305–309
Ashour R, Hamza D, Kadry M, Sabry MA (2023) Molecular detection of Babesia microti in dromedary camels in Egypt. Trop Anim Health Prod 55(2):91
Aslam F, Saleem G, Ashraf K, Hafeez MA, Saqib M (2023) Identification and molecular characterization of Theileria annulata with associated risk factors in naturally infected camels from selected districts in Punjab, Pakistan. Pakistan Vet J 43(1):1614
Alanazi A, Alyousif M, Hassieb M (2012) Seroprevalence study on Theileria equi and Babesia caballi antibodies in horses from central province of Saudi Arabia. J Parasitol 98(5):1015–1017
Ismael AB, Swelum AA, Khalaf AF, Abouheif MA (2014) Clinical, haematological and biochemical alterations associated with an outbreak of theileriosis in dromedaries (Camelus dromedarius) in Saudi Arabia. Pak Vet J 34(2):209–213
Abdallah HR, Faye B (2012) Phenotypic classification of Saudi Arabian camel (Camelus dromedarius) by their body measurements. Emir J Food Agric 24:272–280
Fallatah S, Ghallab E, Khater E (2019) Phylogenetic diversity and DNA barcoding of the camel tick Hyalomma dromedarii (Acari: Ixodidae) of the Eastern region of Saudi Arabia. Trop Biomed 36(2):390–401
El-Hawagry MS, Khalil MW, Sharaf MR, Fadl HH, Aldawood AS (2013) A preliminary study on the insect fauna of Al-Baha Province, Saudi Arabia, with descriptions of two new species. ZooKeys 274:1–88
Bett B, Lindahl J, Delia G (2019) Climate change and infectious livestock diseases: the case of rift valley fever and tick-borne diseases. In: Rosenstock TS, Nowak A, Girvetz E (eds) The climate-smart agriculture papers: investigating the business of a productive, resilient and low emission future. Springer, Cham, pp 29–37
Alanazi AD, Al-Mohammed HI, Alyousif MS, Said AE, Salim B, Abdel-Shafy S, Shaapan RM (2019) Species diversity and seasonal distribution of hard ticks (Acari: Ixodidae) infesting mammalian hosts in various districts of Riyadh Province, Saudi Arabia. J Med Entomol 56(4):1027–1032
ElGhali A, Hassan S (2010) Life cycle of the camel tick Hyalomma dromedarii (Acari: Ixodidae) under field conditions in Northern Sudan. Vet parasitol 174(3–4):305–312
Randolph SE, Asokliene L, Avsic-Zupanc T, Bormane A, Burri C, Gern L, Golovljova I, Hubalek Z, Knap N, Kondrusik M (2008) Variable spikes in tick-borne encephalitis incidence in 2006 independent of variable tick abundance but related to weather. Parasit Vect 1(1):1–18
Mahmoud SH, Mohammad F, Alazba A (2014) Determination of potential runoff coefficient for Al-Baha Region, Saudi Arabia using GIS. Arab J Geosci 7(5):2041–2057
Mahmoud SH, Alazba A (2015) The potential of in situ rainwater harvesting in arid regions: developing a methodology to identify suitable areas using GIS-based decision support system. Arab J Geosci 8(7):5167–5179
Mahmoud SH, Mohammad F, Alazba A (2015) Delineation of potential sites for rainwater harvesting structures using a geographic information system-based decision support system. Hydrol Res 46(4):591–606
Al-Aklabi A, Al-Khulaidi AW, Hussain A, Al-Sagheer N (2016) Main vegetation types and plant species diversity along an altitudinal gradient of Al-Baha region, Saudi Arabia. Saudi J Biol Sci 23(6):687–697
Walker AR (2003) Ticks of domestic animals in Africa: a guide to identification of species (Vol. 74). Biosci Rep
Beati L, Keirans JE (2001) Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J Parasitol 87(1):32–48
Black WC, Piesman J (1994) Phylogeny of hard-and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Pro Natil Acad Sci USA 91:10034–10038
Chandra S, Smith K, Alanazi AD, Alyousif MS, Emery D, Šlapeta J (2019) Rhipicephalus sanguineus sensu lato from dogs and dromedary camels in Riyadh, Saudi Arabia: low prevalence of vector-borne pathogens in dogs detected using multiplexed tandem PCR panel. Folia Parasitol 66:1–13
Folmer O, Hoeh WR, Black MB, Vrijenhoek RC (1994) Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Mol Mar Biol 3(5):294–299
Diab FM, Al-Khalifa MS, Al-Asgah NA, Hussein HS, Khalil GA (2006) Ticks (Acari: Argasidae, Ixodidae) infesting livestock in Saudi Arabia. Fauna Arabia 22:233
Al-Khalifa MS, Al-Asgah NA, Diab FM (1984) Ticks (Acari: Ixodidae) infesting common domestic animals in AL-Qasim province, Saudi Arabia. J Med Entomol 21:114–115
Alanazi A, Al-Mohamed H, Alysousif M, Puschendorf R, Abdel-Shafy S (2018) Ticks (Acari: Ixodidae) infesting domestic and wild mammalians on the Riyadh province, Saudi Arabia. J Entomol 15:1812–5670
Bakheit MA, Latif AA, Vatansever Z, Seitzer U, Ahmed J (2012) The huge risks due to hyalomma ticks. In: Mehlhorn H (ed) Arthropods as vectors of emerging diseases, vol 3. Springer, Berlin
Diab FM, Hoogstraal H, Wassef HY, Al Khalifa MS, Al Asgah NA (1985) Hyalomma (Hyalommina) arabica: nymphal and larval identity and spiny mouse hosts in Saudi Arabia (Acarina: Ixodoidea: Ixodidae). J Parasitol 71:630–634
Telmadarraiy Z, Vatandoost H, Chinikar S, Oshaghi MA, Moradi M, Ardakan EM, Nasiri A (2010) Hard ticks on domestic ruminants and their seasonal population dynamics in Yazd Province. Iran J Arthropod Borne Dis 4(1):66–71
Rahbari S, Nabian S, Shayan P (2007) Primary report on distribution of tick fauna in Iran. Parasitol Res 101:175–177
Nourollahi Fard SR, Fathi S, Norouzi Asl E, Asgary Nazhad H, Salehzadeh Kazeroni S (2012) Hard ticks on one-humped camel (Camelus dromedarius) and their seasonal population dynamics in southeast. Iran Trop Anim Health Prod 44:197–200
Noaman V, Abdigoudarzi M, Nabinejad AR (2017) Abundance, diversity and seasonal dynamics of hard ticks infesting cattle in Isfahan province. Arch Razi Inst 72(1):15–21
Dautel H, Dippel C, Kämmer D, Werkhausen A, Kahl O (2008) Winter activity of Ixodes ricinus in a Berlin forest. Int J Med Microbiol 298:50–54
Abdullah HH, El-Shanawany EE, Abdel-Shafy S, Abou-Zeina HA, Abdel-Rahman EH (2018) Molecular and immunological characterization of Hyalomma dromedarii and Hyalomma excavatum (Acari: Ixodidae) vectors of Q fever in camels. Vet World 11(8):1109
Shubber HWK, Al-Hassani NAW, Kadhim M (2014) Research article ixodid ticks diversity in the middle and South of Iraq. Int J Recent Sci Res 5(9):1518–1523
Apanaskevich DA, Schuster AL, Horak IG (2008) The genus Hyalomma: VII. Redescription of all parasitic stages of H. (Euhyalomma) dromedarii and H.(E.) schulzei (Acari: Ixodidae). J Med Entomol 45(5):817–831
Oorebeek M, Kleindorfer S (2008) Climate or host availability: what determines the seasonal abundance of ticks? Parasitol Res 103(4):871–875
Al-Deeb MA, Muzaffar SB, Abu-Zeid YA, Enan MR, Karim S (2015) First record of a spotted fever group Rickettsia sp. and Theileria annulata in Hyalomma dromedarii (Acari: Ixodidae) ticks in the United Arab Emirates. Fla Entomol 98(1):135–139
Telmadarraiy Z, Vatandoost H, Rafinejad J, Mohebali M, Tavakoli M, Abdigoudarzi M, Salari SER (2009) Distribution of ticks (Ixodidae and Argasidae) family and susceptibility level to cypermethrin in Meshkinshahr district, Ardabil province, Iran. Ardabil Uni Med Sci J 9(2):127–133
Durrani AZ, Shakoori AR, Kamal N (2008) Bionomics of Hyalomma ticks in three districts of Punjab. Pak J Anim Plant Sci 18(1):17–23
McCartan B, Hunter A, Pegram R, Bourne A (1987) Tick infestations on livestock in the Yemen Arab Republic and their potential as vectors of livestock diseases. Trop Anim Health Prod 19(1):21–31
Pegram RG, Hoogstraal H, Wassef HY (1982) Ticks (Acari: Ixodoidea) of the Yemen Arab Republic. I. Species infesting livestock. Bull entomol Res 72(2):215–227
Al Thabiani A, Panneerselvam C, Alshehri MA, Asiry KA, Alsaif M, Alhowity Y (2021) Efficacy of synthetic pyrethroids on camel ticks Hyalomma dromedarii “Acari: Ixodidae” in Saudi Arabia. Entomol Appl Sci Lett 8:27–32
Abdally M (2008) Identification of hard tick species affecting camels (Camelus Dromedarius) and their seasonal abundance in Najran, Saudi Arabia. Alexa Sci Exch J 29:71–76
Apanaskevich DA, Horak IG (2008) The genus Hyalomma Koch, 1844: V. Re-evaluation of the taxonomic rank of taxa comprising the H. (Euhyalomma) marginatum Koch complex of species (Acari: Ixodidae) with redescription of all stages and notes on biology. Int J Acarol 34(1):13–42
Al-Deeb MA, Enan MR (2018) Genetic diversity in the camel tick hyalomma dromedarii (Acari: Ixodidae) based on mitochondrial cytochrome c oxidase subunit I (COI) and randomly amplified polymorphic DNA polymerase chain reaction (RAPD-PCR). Advs Entomol 6(4):285–294
Lv J, Wu S, Zhang Y, Chen Y, Feng C, Yuan X, Jia G, Deng J, Wang C, Wang Q (2014) Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida). Parasit Vect 7(1):1–11
Al-Khalifa MS, Diab FM, Al-Asgah NA S (1983). A checklist of ticks (Ixodoidea) infesting local farm animals in Saudi Arabia. I. Ticks of Al-Qasim region
Alanazi AD, Nguyen VL, Alyousif MS, Manoj RR, Alouffi AS, Donato R, Sazmand A, Mendoza-Roldan JA, Dantas-Torres F, Otranto D (2020) Ticks and associated pathogens in camels (Camelus dromedarius) from Riyadh, Province Saudi Arabia. Parasit Vect 13(1):1–9
Omer SA, Alsuwaid DF, Mohammed OB (2021) Molecular characterization of ticks and tick-borne piroplasms from cattle and camel in Hofuf, eastern Saudi Arabia. Saudi J Biol Sci 28(3):2023–2028
Pegram RG, Higgins AJ (1992) Camel ectoparasites: a review. In: Proceedings of the first international camel conference, Dubai, UAE, pp 2–6
Acknowledgements
The authors thank the Deanship of Scientific Research et al. Baha University for providing support in conducting this study (8/1442).
Funding
This study was funded by Department of Biology, Faculty of Science, Al-Baha University, 8/1442, Samia Alghamdi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declare that they have no conflict of interest.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alghamdi, S.Q. Prevalence of tick infestation and molecular characterization of tick species of camels (Camelus dormedarius) from Al-Baha South Saudi Arabia. J.Umm Al-Qura Univ. Appll. Sci. (2024). https://doi.org/10.1007/s43994-024-00138-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43994-024-00138-w