Abstract
Cholera infection results from the ingestion of water or food contaminated with toxigenic Vibrio cholerae. This study evaluated the occurrence of toxigenic V. cholerae in Asata River, Enugu Metropolis, Nigeria, and estimated V. cholerae infection risks from use of the River water for drinking, domestic and recreational purposes. Vibrio was detected and quantified using membrane filtration technique and thiosulfate–citrate–bile salts–sucrose agar. Isolates were screened by PCR, using specific primers targeting the internal transcribed spacer (its) region between 16 and 23S rRNA and the cholera toxin (ctx) gene. Sequenced 16SrRNA gene amplicons of two selected isolates were used for phylogenetic analysis. Quantitative microbial risk assessment (QMRA) was conducted using the β-Poisson dose–response model. About 81% (58/72) of Asata River samples recorded Vibrio counts above 1.0 \(\times\) 103 cfu/100 mL. Of the fifty Vibrio isolates screened, its was detected in 54% (27/50), out of which 74% (20/27) had the ctx gene of toxigenic V. cholerae. Evolutionary relatedness of the sequenced isolates to V. cholerae was revealed. The estimated risks of cholera infection in persons exposed to untreated Asata River water were above 0.5 for all the exposure scenarios, for both the rainy and dry seasons. The risks were highest (~ 0.9) for exposure via drinking water and annual risk of infection was deduced to have a probability of 1.0. Therefore, dependence on the untreated Asata River water for drinking, recreational, domestic and irrigation purposes may present a potential public health risk of cholera outbreak. We recommend increased monitoring and surveillance of River water for Vibrio abundance and that Asata River be protected from further degradation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Rivers and streams remain a vital source of water for drinking, household, and other purposes in many developing countries. Globally, more than 144 million people depend directly on surface waters for drinking water, with 58% of these people living in sub-Saharan Africa [1]. Poor sanitation practices in countries in the region have the potential to contaminate surface waters with faeces originating from both human and animal sources. In 2019, UNICEF and WHO estimated that about 2 billion people lack basic sanitation and that about 673 million people practice open defecation worldwide. Notably, Nigeria is among the countries with a high rate of open defecation, with only about 20% of the population, in both urban and rural areas, using water that is free from faecal contamination [1]. Further, over 80% of wastewater from human activities—sewage treatment plants, slaughterhouses, livestock farms and household’s wastewater are discharged directly into surface waters with minimal pre-treatment [2]. Combination of all these poor practices makes surface freshwater bodies a thriving habitat for waterborne pathogens that cause enteric diseases such as diarrhoea, cholera, and other gastrointestinal diseases a major public health concern [3].
Cholera, which is characterized by acute episodes of diarrhoea can occur after ingestion of toxigenic V. cholerae. V. cholerae strains that possess the cholera toxin gene ctx are toxigenic [4]. Toxigenic V. cholerae strains produce enterotoxin, which contributes to its pathogenicity [4]. When the V. cholerae escapes the stomach gastric juice and colonizes the small intestinal mucosa, it produces a potent enterotoxin (cholera toxin, or CTX). CTX binds to the intestinal walls and disrupts the normal flow of sodium and chloride ions, resulting in the excretion of enormous amounts of fluid leading to diarrhoea, water and salt (electrolyte) loss. Diarrhoea is the leading cause of death in people of all ages and the fourth leading cause of death in children under the age of five [5]. Further, it is estimated that 3 to 5 million cases of cholera occur worldwide with approximately 100,000 to 143,000 deaths annually [6]. According to the World Health Organization (WHO), sub-Saharan Africa carries the largest burden of cholera, with Nigeria among the highest risk countries. Estimates suggest that there are more than a hundred thousand cholera patients in Nigeria, with about 220,000 cases and 8000 deaths predicted each year [5].
Cholera infection can be asymptomatic or symptomatic with moderate, mild, or severe cases. In some cases, cholera results in bacteraemia depending on the patients’ predisposing factors such as the presence of chronic medical condition [7], blood type [8], gastric acidity, and hygienic condition of the infected person [9]. In vulnerable individuals, the rapid loss of body fluids can lead to dehydration and shock, and without treatment, death can occur within hours [10]. In asymptomatic individuals, the bacterium may remain in the faeces for 1–10 days following infection [11]. In such cases, bacterial cells can be released back into the environment to potentially infect healthy people if proper hygienic practices are not followed. About 20% of infected individuals will have severe symptoms of the disease characterized by profuse watery diarrhoea (with extreme fluid and electrolyte depletion, and severe dehydration), vomiting, and leg cramps [11, 12]. The risk of cholera infection is highest among the poor, the most vulnerable population groups in developing countries, particularly in Africa, Asia, and some parts of North and Central America [13], where access to safe drinking water and adequate sanitation is a major challenge. Thus, cholera is regarded as a disease of inequity [11].
In Nigeria, cholera is endemic, occurring annually, albeit mainly during the rainy season and mostly in areas with insufficient access to drinking water and poor sanitation [12]. This occurs because rainwater carries waste into drains and subsequently into surface waters. Intense rainfall during rainy season can lead to overflow of sewage system, thereby compromising overall sanitation standards [12]. Increased risks of cholera and other water-borne infections has been attributed to climate change, stemming from rising global temperatures and sea levels, combined with irregular rainfall patterns leading to more frequent flooding [13]. Floods carry infectious agents from farmlands and slaughterhouses to water bodies.
The first series of cholera outbreaks in Nigeria were reported between 1970 and 1971 [14]. Major epidemics also occurred in 1992, 1995–1996 and 1997. The Federal Ministry of Health in Nigeria reported 37,289 cases and 1434 deaths between January and October 2010, while a total of 22,797 cases of cholera with 728 deaths and a case-fatality rate of 3.2% were recorded in 2011. Outbreaks were also recorded in 2018 with the Nigeria Centre for Disease Control (NCDC) reporting 42,466 suspected cases including 830 deaths from 20 of the 36 States in Nigeria. In addition, the 2021 cholera outbreak in Nigeria affected 34 of the 36 states with a fatality rate of 3.2%. By the end of 2021, the NCDC recorded approximately 111,000 suspected cases with 3604 suspected deaths from cholera, a number that is higher than the total number of COVID-19 deaths reported in Nigeria as of January 2022 [15]. There is currently a lack of data on cholera cases for health facilities in Enugu. Nonetheless, an epidemiological study focusing on Nigeria’s cholera outbreak in 2020–2021 estimated that there were about 127 cholera cases in Enugu, accompanied by a remarkable 10% fatality rate (CFR) [16].
An abundance of V. cholerae in freshwater bodies has been reported to precede cholera outbreaks [17]. V. cholerae has approximately 200 serotypes based on the structure of the lipopolysaccharide (LPS) cell wall ‘O’ antigen. However, only two serotypes, O1 and O139, have been associated with large scale, cross-boundary cholera epidemic/pandemic. All other serotypes, referred to as non-O1 and non-O139, as well as the non-toxigenic O1 and O139 serotypes, can cause gastroenteritis or cholera-like diarrhoea and bacteraemia [7, 20, 21]. Pathogenic V. cholerae strains are distinguished from non-pathogenic ones by the presence of specific virulence genes such as tcp, zot, tox, hly, tcol, and especially, the ctx gene [4] that can be transferred horizontally between strains [22]. Strains that cause cholera possess a filamentous bacteriophage (ctxΦ) gene that encodes the cholera toxin protein. In addition, pathogenic strains of V. cholerae may possess a virulent factor-toxin co-regulated pilus (TCP) protein, which is essential for colonization and may also serve as a receptor for the CTXΦ protein. V. cholerae O139 was first identified in Bangladesh in 1992 [21]. The serotype, O139 are mostly restricted to South-eastern Asia and is responsible for many outbreaks in Bangladesh [21]. However, reports from several studies on cholera outbreaks suggest that ctx-positive V. cholerae O1 is responsible for most of the recent cholera epidemics around the world.
Some studies have reported the occurrence of V. cholerae in water sources and aquaculture [23,24,25,26,27,28,29,30] in outbreak and non-outbreak cases in Nigeria. Other studies linked cases of cholera outbreak to the use of surface water [31, 32]. However, limited studies in Nigeria have quantitatively detected Vibrio in a river water over time and space nor, conducted risk assessment of exposure to surface water contaminated with V. cholerae. In this study, we investigated the incidence of toxigenic V. cholerae in water samples collected from the Asata River, in Enugu Metropolis, Nigeria. The overarching objective of the study was to quantitatively assess the public health risk of cholera infection from exposure to the river water.
2 Methodology
2.1 Study area/site description and sample collection
The study area consists of the three Local Government Areas; Enugu North, Enugu South and Enugu East that form the Enugu Metropolis, the capital of Enugu State, Nigeria. Asata River is one of the two major rivers that flow through the Metropolis and six residential areas that make-up the Enugu urban region. It is a perennial river spanning 19.8 km and having Akwata and Idaw rivers, as well as a few streams as its tributaries [33]. Asata River is heavily impacted by anthropogenic activities such as unsafe disposal of untreated sewage, wastewater effluent, open defecation, indiscriminate dumping of household and commercial waste, herd watering, direct disposal of slaughterhouse effluent, and fertilizer pollution from riverbank farming. Due to unavailability of piped water supply in Enugu Metropolis, which has an estimated population of about 816,000, a significant number of the residents use Asata River water for drinking, domestic, irrigation and recreational purposes [34].
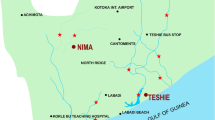
On a monthly basis, water samples were collected from six selected sampling sites, S1 to S6 (Fig. 1) along the river course, for a sampling period of one year (September 2017 to August 2018). That is, 6 water samples from six sites, every month for 12 months, giving a total of 72 samples. March to October was designated as the rainy season and November to February the dry season, based on deductions from meteorological data of the study area [34]. The sampling sites were chosen based on high rates of anthropogenic activity as well as the availability of access to the river, large water usage, and density of human settlement. The geo-coordinates of the selected sites, determined using a GPS navigator, are given as footnotes to the Google Earth map (Fig. 1).
Map showing the six sampling sites (S1–S6) on Asata River. [**S1 (Recreation Point): 6°26′2.977″N; 7°27′50.435″E, S2 (Coal Camp Bridge): 6°25′59.303″N; 7°28′30.935″E, S3 (Zik’s Avenue Bridge): 6°26′10.248″N; 7°29′40.092″E, S4 (Presidential Road): 6°26′34.513″N; 7°30′4.752″E, S5 (EN-PH Road Bridge): 6°27′18.36″N; 7°32′23.136″E, S6 (Before Ekulu-Asata Confluence): 6°44′30.92″N; 7°50′14.21″E]
Water samples were collected from each of the six sampling sites using sterile 500-ml screw-capped Pyrex glass bottles. The collection procedure followed the standard practice for surface water collection, dipping the sterile bottle below 5 cm and facing upstream, against the flow of the river. The collected water samples were transported to the Water and Public Health Research Laboratory, University of Nigeria, Nsukka in an insulation box and analysed in triplicates for the presence of Vibrio within 4 h of sample collection.
2.2 Isolation and quantitative evaluation of Vibrio species
Detection and quantification of Vibrio species were carried out using membrane filtration as previously described [35]. Briefly, a 100-ml dilution of each water sample was filtered through a sterile 0.45 µm pore size membrane filter (Merck: Darmstadt, Germany). The filters were aseptically transferred to sterile thiosulfate-citrate-bile-salts-sucrose (TCBS) agar medium (Oxoid: Hampshire, UK) and incubated at 35 °C for 24 h. After incubation, the presumptive Vibrio colonies (appearing as green and yellow colonies, with 2–3 mm in diameter) were counted and cell density was calculated and expressed as colony forming units per 100 ml (cfu/100 ml).
The presumptive Vibrio species were isolated and purified using the method described by Nongogo and Okoh [36] with slight modifications. Briefly, 2–3 discrete presumptive V. cholerae colonies (yellow 2–3 mm in diameter), representing each sampling site per month, were randomly selected and purified by picking from the agar plate and sub-culturing onto sterile TCBS agar plates. After 24 h of incubation at 35 °C, pure cultures were incubated in tryptone soy broth, TSB (LAB-M: Lancashire, UK) at 35 °C for 24 h. The resulting colonies were then stored in 15% sterile glycerol at − 6 °C until further analysis. Preliminary identification was made by macroscopic examination, Gram staining, microscopic and biochemical (oxidase test) analyses. V. furnissii DSM 19622 and V. harveyi DSM 19623 were used as control strains. One-way ANOVA and t-test were used to evaluate if differences in Vibrio concentrations between sites and seasons were statistically significant.
2.3 Molecular identification of Vibrio species
2.3.1 Sanger sequencing of 16 s rRNA
Two pure isolates designated as WZBAV01 and WZBAV02 were sent to Inqaba Biotechnical Industries (Pty) Ltd, South Africa for sequencing. First, to confirm the detection of V. cholerae in Asata River water samples, and second, to determine the relatedness of Asata River isolates to strains implicated in recurrent cholera outbreaks in Nigeria. Genomic DNA was extracted and purified by the Boiling method [36, 37]. The PCR cocktail consisted of 10 µl of 5 \(\times\) GoTaq colourless reaction, 1 µl of 10 mM of dNTPs mix, 3 µl of 25 mM MgCl2, 1 µl of 10 pmol each of the 16S rRNA gene forward (16SF: GTGCCAGCAGCCGCGCTAA) and reverse (16SR: AGACCCGGGAACGTATTCAC) universal primers, and 0.3 units of Taq DNA polymerase (Promega, USA) and 8 μl DNA template. PCR reaction mixtures were made up to 50 µl with sterile distilled water. PCR was carried out in a GeneAmp 9700 PCR System Thermocycler (Applied Biosystem Inc., USA) with a cycle consisting of an initial denaturation at 94 ℃ for 5 min; followed by 30 cycles consisting of 94 ℃ for 30 s, annealing was set at 56 ℃ and 72 ℃ for 30 s and 1 min 30 s respectively. Cycle termination was at 72 °C for 10 min and then, chill at 4 ℃. The PCR amplicons were resolved by gel electrophoresis using 1.5% (w/v) agarose and 1 \(\times\) TAE buffer (40 mM Tris–acetate in 1 mM EDTA, pH 8.0). The gel was pre-stained with 3 µl of 0.5 g/ml ethidium bromide and electrophoresed at 120 V for 45 min and visualized by ultraviolet trans-illumination. Thereafter, the amplified fragments were ethanol purified to remove the PCR reagents.
The purified amplicons were sequenced using Applied Biosystems Genetic Analyzer 3130xl sequencer according to the manufacturers’ protocol. The resulting sequences were used as reference to identify similar/reference 16S rRNA gene sequences using the Basic Local Alignment Tool (BLAST) on the National Center for Biotechnology Information (NCBI) database. Similar 16S rRNA sequences were aligned using the Cluster W tool on the EMBL/GenBank databases. The phylogenetic tree was constructed using MEGA 11 software. The aligned 16S rRNA gene sequences of the samples with the reference sequences retrieved from the NCBI database were used to construct the phylogenetic tree using the maximum likelihood-based phylogenetic tree and bootstrap values (≥ 50%).
2.4 Molecular characterization and detection of virulence gene
A total of 60 isolates (representing all sampling sites) were sub-cultured onto nutrient agar (LAB-M: Lancashire, UK) and incubated at 35 °C for ~ 18 h and subjected to genomic DNA extraction using the boiling method [36, 37]. Discrete colonies (10 per culture plate) from the 18-h overnight cultures were picked and suspended in 200 µl of pyrogen-free sterile water in 2-ml Eppendorf tubes and vortexed vigorously. The suspended cells were lysed by boiling in Eppendorf heating block thermostat 5320 (Eppendorf Geratebau Netheler, HinzGmbH, Germany) for 10 min at 100 °C. The cells were then chilled on ice for 5 min and centrifuged at 3913×g. The DNA-containing supernatant was transferred to a fresh sterile Eppendorf tube. The quality of the extracted DNA was then determined using Epoch Nanodrop (BioTek Instruments Inc., Winooski, VT, USA) at 260 and 280 nm. DNA samples with 260/280 nm range of 1.80–2.20 were used as template for the PCR-based assay. A total of fifty DNA samples met the criteria and were screened by PCR using primers specific for the internal transcribed spacer (its) region between 16 and 23S rRNA, and ctx were used to identify V. cholerae and toxigenic V. cholerae, respectively. The target amplicons (with the respective sizes) and primer sequences are listed in Table 1.
PCR was performed in a C1000 Touch™ Thermal Cycler equipped with CFX96™ Deep Well Reaction Module (Bio-Rad, Hercules, CA, USA). The PCR reaction mixture contained genomic DNA (5 ng/µl), specific primers (0.5 µM each), dNTPs (0.1.25 mM) and PrimeStar® GXL DNA polymerase (1 µl; Clontech-Takara, Mountain View, CA) in a 50 µL reaction mixture. The thermocycling conditions were as follows: initial denaturation (98 °C for 2 min in 1 cycle), denaturation (98 °C for 20 s), annealing (annealing temperature was set between 50 and 58 °C) for 30 s, extension (72 °C for 1 min in 35 cycles), final extension (72 °C for 10 min in 1 cycle), and hold (4 °C for 10 min in 1 cycle). The resulting amplicons were resolved by gel electrophoresis using agarose (1.5%, w/v) and 1 \(\times\) TAE buffer (40 mM Tris–acetate in 1 mM EDTA, pH 8.0). Gel electrophoresis was conducted at 125 V and 100 mA for 30 min using EC1000XL programmable electrophoresis power supply (Thermo Scientific, Waltham, MA, USA). The agarose gel was pre-stained with ethidium bromide (0.2 µg) and the images were viewed under UV (ultraviolet) light using Bio-Rad Chemi-Doc Imager (BioRad, Hercules, CA). The bands and sizes were determined from a 1 Kb DNA ladder as a reference.
2.5 Risk assessment [quantitative microbial risk assessment (QMRA)]
QMRA estimates the possibility of an adverse human health effect after contact or exposure with a particular pathogen. It involves a four steps approach: hazard identification, exposure assessment, dose–response relationship, and risk estimation [37, 40, 41].
2.5.1 Hazard identification
Pathogenic V. cholerae strains possess virulence determinants, particularly the cholera toxin gene (ctx). V. cholerae is transmitted through faecal-oral route via ingestion of contaminated water or food, and to lesser extent, through direct contact with an infected person [42]. Although there are reports of transmission through skin cuts, wounds, ears, etc., it must be emphasized that these routes of transmission in the environment are rare [43]. The infectious dose can be as low as 1 \(\times\) 103 in people with reduced gastric acidity [4], and the disease endpoint, cholera, can be mild, moderate, or severe, which can quickly lead to death if not promptly treated [10]. The disease can lead to epidemic and pandemic proportions, and the burden is greater in developing countries that lack adequate safe drinking water and sanitation [13]. In Nigeria, outbreaks are reported annually in different states with a national case fatality rate (CFR) of ~ 3.0% [14, 17].
2.5.2 Exposure assessment
In this study, exposure assessment was based on three variables; mean concentration of Vibrio counts obtained from each of the six Asata River sampling sites (X in cfu/100 ml), the percentage of confirmed toxigenic V. cholerae (ctx-positive) strains from our PCR-based assay (Y%), and an assumed volume of ingested water via different exposure pathways (W in mL). The variables were used to calculate the dose (d) of toxigenic V. cholerae ingested via each exposure route per sampling site using a derived formula (Eq. 1) below:
The values and result outputs are shown in Table 2.
2.5.2.1 Assumptions
Multiple exposure scenarios considered for ingestion of Asata River water include drinking, accidental ingestion of water during bathing and other recreational activities or while using of the water for household purposes such as dishwashing or laundry, and irrigation and the associated consumption of contaminated food. Thus, exposure to V. cholerae infection can occur via multiple pathways. For example, a person may drink water from the Asata River and consume vegetables from a farm that used the Asata River water for irrigation. The amount of water consumed varies by individual, culture, and climate. We assumed that the volume of water consumed/day via different exposure scenarios were 1000 ml, 30 ml, 10 ml, and 2 ml for persons exposure by drinking untreated Asata River water, and through recreational, domestic, and irrigation purposes respectively [41, 44].
2.5.3 Dose–response analysis and risk characterization
The dose–response estimates the cholera risk from the dose of toxigenic V. cholerae ingested via each exposure route per sampling site, while the risk characterization determines the magnitude of the risk with or without reference to uncertainties. In this study, risk characterization integrates the dose (exposure assessment) with risk at different doses (dose–response) to estimate a probability of infection. The risk of infection/day (with toxigenic V. cholerae was estimated using the β-Poisson model (Eq. 2) which provides a suitable fit to the available data set.
Where,
Pinfection/day = Probability of becoming infected by ingesting dose (d) number of the bacterial pathogens
d = Dose of infectious bacterial cells ingested (Eq. 1)
N50 = Median infectious dose, indicating the number of organisms that will infect 50% of the exposed population (a known median infectious dose for V. cholerae was used)
α = β-Poisson model dose-response constant
Previously published dose–response parameters were used, and the reported α and N50 values for V. cholerae are 0.2500 and 243 respectively [45]. One of the main flaws of QMRA is the subjectivity in model and parameter selection [45].
2.5.4 Annual risk of infection
The frequency of Asata River water ingestion and Asata River–irrigated crops consumption is important in determining the annual risk of infection. Therefore, this study computed the annual risk of infection (Pinfection/year) for V. cholerae as a function of daily risk using Eq. 3. A risk of more than 1 in 10,000 people per year was considered an unacceptable risk of infection.
2.5.5 Sensitivity analysis
Sensitivity analysis was performed, following a deterministic approach [46], to investigate the contribution of dose–response parameter to the output of the risk model. For deterministic models and for a crude sensitivity screening in probabilistic models, the nominal range sensitivity analysis (NRSA) is appropriate in many cases. In NRSA, the value of one model input parameter is varied over its range of plausible values while all other model inputs are kept at their baseline value, and the magnitude of change of the calculated model output is recorded [44]. In this basic method for sensitivity analysis, varied the dose–response parameter for β-Poisson model, to determine the magnitude of change of the estimated risk. We used the α values 0.01, 0.05, 0.15, 0.25, 0.35, 0.45, and 0.55, and this sensitivity analysis applies to the annual risk of infection, morbidity and mortality as they are all functions of the daily risk of infection. The probability of clinical illness was determined by multiplying the daily risks of infection by 0.5, while the probability of mortality was determined by multiplying the probability of illness by 0.01% for the general population [41].
The occurrence of Vibrio in surface waters is known to be affected by location and season. Two representative sites (S1 and S2) and the two seasons (Rainy and Dry), all of which recorded the detection of the toxigenic strain of this pathogen, were chosen to determine the influence of dose–response values on the daily risks of V. cholerae infection. S1 was selected because it is located in the upper catchment of the Asata River with reduced human activity and water is abstracted from this point into reservoirs and, without treatment, distributed by water tankers to residents on Enugu metropolis, while S2 has both urban and rural communities in its catchment and the highest mean Vibrio count was detected at this site. Also, site S3 was chosen because it recorded the biggest (3.4-fold) higher mean Vibrio count during the rainy season than the dry season, and changed the input dose–response parameter values to determine the magnitude of change of the estimated risks in the two seasons. Opting for the worst-case scenario, drinking water use was chosen as it results in the consumption of significantly higher volume of water, compared to other exposure scenarios.
3 Results
3.1 Quantitative detection and confirmation of V. cholerae
Vibrio species were widely distribution across the sampling sites and sampling months. The lowest counts were recorded at S1, a location near the mouth of the river. The S1 site is characterized with lower human presence and have little to no observable anthropogenic activity. Conversely, Vibrio load at site S2 was significantly higher than at all other sites. The overall total Vibrio counts in the study river ranged from 1.0 \(\times\) 100 to 8.0 \(\times\) 105 cfu/100 ml (Table 2). Although we recorded mean differences between total Vibrio counts at different sampling sites during the rainy and the dry season (Table 3), the counts were not significantly different except at site S3, and for mean value of all river water sample collected per season (Table 3).
The sequences of isolates WZBAV_01 and WZBAV_02 (deposited with the NCBI accession numbers OR435834 and OR435835, respectively) showed evolutional relatedness to V. cholerae and V. fluvalis, respectively. The phylogenetic relatedness using the Neighbour-joining method is illustrated in Figs. 2 and 3.
Detection of its has been used as possible confirmation of V. cholerae, whereas the detection of ctx indicate toxigenic strains of V. cholerae [4]. Of the fifty isolates screened for both genes, its was detected in 54% (27/50), out of which 74% (20/27) had the ctx gene of toxigenic V. cholerae. Toxigenic V. cholerae (ctx-positive) isolates in this study, accounts for 40% (20/50) of the screened isolates.
3.2 Risk assessment
The mean Vibrio counts at each sampling site were used to estimate the concentration of toxigenic V. cholerae (based on 40% toxigenic V. cholerae (ctx-positive) detected by PCR) per 100 ml water sample. An estimated concentration of toxigenic Vibrio, 137 and 71 cfu/100 ml was recorded at (S1) during the rainy and dry seasons, respectively (Table 4). The corrected infectious dose was calculated for the different routes of exposure as shown in Table 4. Using the β-Poisson dose–response model, it was found that the probability of infection/day after ingestion of the Asata River water was above 0.5 during both the rainy and dry seasons, (Table 5) for all possible exposure scenarios—drinking, recreational, domestic, and irrigation uses. As expected, the daily risk of cholera infection from drinking water from the Asata River was the highest (~ 0.9), at all sampling sites during the raining and dry seasons except at S1 with a daily risk of ~ 0.7 (Table 5). The annual risk of infection was deduced to have a probability of 1.0 for all exposure scenarios (Table 5). The results from the sensitivity analysis simulations are presented in probability plot format in Fig. 4.
4 Discussion
There are increasing reports of Vibrio abundance in water bodies across the world and the threat of vibriosis is becoming more common in both developing and developed countries [7]. In this study, we investigated the distribution of Vibrio species in the Asata River in Enugu, South-Eastern Nigeria during both the rainy and dry seasons for a 12-month sampling period. Our data showed that Vibrio species were widespread during the sampling months and across the sampling sites. The abundance of Vibrio in water bodies has been shown to precede cholera outbreaks [19]. For the Vibrio counts recorded in this study, 80.5% (58/72) of the samples contained more than 1000 cfu/100 ml. This is a major concern as the infectious dose of V. cholerae has been shown to be as low as 1.0 \(\times\) 103 cfu [4]. As our climate continues to change, there are projections of increased temperature and rainfall patterns. This might give rise to the spread of V. cholerae in surface water thereby, increasing the risk of disease.
The high mean Vibrio counts (2.8 × 102 to 1.2 × 105) reported in this study (Table 2) could be attributed to the warm surface water temperatures > 24 ℃ of the Asata River [33, 34]. Vibrio species have been reported to be more abundant in surface waters with temperatures above 15 °C with maximum pathogenic vibrios counts, including V. cholerae and V. parahaemolyticus obtained in surface waters > 25 ℃ [19, 35, 47]. In addition to the water temperature, the impact of anthropogenic activities evident at the sampling sites could also be responsible for the Asata River’s high Vibrio load and the significantly varying counts of Vibrio evident at the sampling sites. Anthropogenic activities were more prominent at site S2, hence the significantly higher count at that site. In this study area, there are ample evidences of faecal pollution from open defecation, leaking septic tanks, animal waste from slaughterhouses and livestock market. In a case in Kasesse District in Uganda, cholera outbreak was traced to an index case who defecated near a community water collection site [48]. We speculate that these activities may be responsible for the high counts of Vibrio species at the sample sites.
Asata River receives a variety of solid and liquid wastes from households, slaughterhouses, and a livestock market, as well as leaking and dilapidated septic tanks. A high prevalence of Vibrio species including antibiotic resistant strains have been reported for surface waters impacted by wastewater, cow dungs, and slaughterhouses effluents in sub-Saharan Africa [25, 27, 49, 50]. Taken together, anthropogenic activities in and around Asata River likely account for the prevalence and abundance of Vibrio in this River. It is worth noting, however, that the quantification method used in each analysis can affect the counts of Vibrio. In this study we used the membrane filtration (MF) method, and in a study in which three cultural methods: MF, direct plating, and most probable number (MPN) were used to determine the prevalence and abundance of Vibrio species in freshwater/brackish surface water bodies, MF appeared to be the most sensitive and reliable method. Particularly, the MF method was able to detect V. cholerae up to 3900 cfu/100 ml in 22 out of 36 water samples, while MPN and direct plating failed to detect V. cholerae in up to 15 and 16 water samples, respectively [51].
The two sequenced isolates, WZBAV_01 and WZBAV_02, showed an evolutionary relatedness to V. cholera and V. fluvialis, respectively. The confirmation of cultural isolates of Vibrio by PCR assay most often detects V. fluvialis as one of the identified species [49, 50, 52]. This could be because V. fluvialis shares a similar phenotypic and genomic similarity with V. cholerae [52]. While V. cholerae is known to cause cholera, an endemic disease in Nigeria and other developing countries [13], V. fluvialis, on the other hand, is recognized as an emerging waterborne and foodborne pathogen that can cause gastroenteritis, septicemia, and severe diarrhoea [50, 52]. Interestingly, it was found that 54% (27/50) of the isolates from this study are V. cholerae (its-positive isolates). This result underscores the importance of molecular tools to characterize isolates. It is important to emphasize that 74% (20/27) of the confirmed V. cholerae strains isolated in this study had the cholera toxin (ctx) gene. This finding is of concern as the presence of the ctx gene in V. cholerae indicates the ability of the strain to cause an infection.
The detection of V. cholerae strains containing the ctx gene could provide a possible explanation for the endemicity of cholera in the area and the recurrent outbreaks in and around Enugu [17]. It could further be inferred that toxigenic strains of V. cholerae are abundant among the human population in the study area, many of whom might be asymptomatic carriers. Asymptomatic infections account for about 80% of cholera cases [11]. Many documented cholera outbreaks have been attributed to contamination of surface water with faecal matter, mostly from asymptomatic individuals [31, 32]. Considering the amount of human and animal waste that enters the Asata River throughout the year, the percentage toxigenic V. cholerae (74%) in this study is not surprising. Furthermore, the significantly lower occurrence of V. cholerae and toxigenic V. cholerae in water samples from site S1—which is largely devoid of anthropogenic activities—implicates human activities, particularly waste disposal into the Asata River as a major driver of the abundance V. cholerae in areas of the Asata River with high human activities.
We observed that the abundance of Vibrio in the water samples collected from Asata River is dependent on season, with the rainy season recording a mean Vibrio count (5.3 \(\times\) 104a ± 8.7 \(\times\) 103) that was significantly higher than that of the dry season (3.3 \(\times\) 104b ± 5.6 \(\times\) 103) as revealed in Table 3. Except for S4 (where the Vibrio counts remained unchanged in both season), all the sampling sites, recorded rainy season counts that ranged from 1.5-fold to 3.4-fold higher Vibrio counts during the rainy season compared to counts observed during the dry season. This could be attributable to the usually higher temperatures of the rainy season as previously reported [34]. Previous studies [19, 35] have reported this dependence on season, but the unexpected observation at site S4 might be due to the fact that it is located in a densely populated urban slum where untreated sewage, effluents and waste are continuously discharged into the river, resulting in a high propensity to introduce faecal matter and possibly Vibrio species into the Asata River.
The results presented in this study show that Asata River water harbours pathogenic strains of V. cholerae implicated in human and animal diseases. Our QMRA data show that the risk of cholera infection was high for all exposure scenarios considered and at all sites sampled during both rainy and dry seasons. In addition, our estimates suggest that drinking Asata River water is the route with the highest probability of infection when compared to other scenarios (Table 5). Due to acute shortage of drinking water, some households, particularly those of low socioeconomic status within the study area commonly use surface Asata River water for drinking, and for domestic and other purposes. This unwholesome practice will further buttress the probability of infection via the exposure routes. This situation and our findings suggest an impending risk of cholera outbreak in Enugu metropolis. With the high probabilities estimated, it would be anticipated that cholera outbreak could take place among populations that depend on the River as a source of water. Incidentally, towards the middle of the year, 2021, a cholera outbreak occurred in communities bordering the Asata River in Enugu metropolis infecting nineteen people and leading to seven deaths [17, 18].
5 Conclusion
In this study, V. cholerae was detected in samples from the Asata River in Enugu metropolis, Nigeria. Although no serological analysis of the isolated V. cholerae was performed, the cholera toxin gene, ctx was detected in 40% of the isolates by PCR. The calculated daily risk of cholera infection from exposure to untreated Asata River water was greater than 0.5 for most exposure scenarios—drinking water, recreation, domestic and irrigation. The risk was highest with a probability of daily infection risk of ~ 0.9 for exposure via drinking water. In addition, the annual risk of infection for all considered exposures during the rainy and dry season was deduced to have a probability of ~ 1.0. Therefore, there is a pressing need for household water treatment and proper sterilization of fresh vegetables and fruits before consumption. Taken together, the Asata River poses a significant health risk to the surrounding communities, particularly given the lack of potable water in the Enugu metropolis. The risk is compounded by the fact that the Asata River also serves as an irrigation source for growing fresh produce. This increases the risk of infection, as fresh produce from the areas around the Asata River can be sold and consumed several miles from where it is grown. A study of the occurrence and density of Vibrio species in vegetables and fruits grown with water from the Asata River and tracing the distribution (in the market) of agricultural produce grown with water from the River will prove instructive towards delineating the role that this River plays in the incidence and spread of cholera in the study area and beyond.
6 Limitations of the study
This study investigated the incidence of pathogenic V. cholerae in water samples collected from the Asata River, an urban river in Enugu Metropolis Region in Nigeria and quantified the microbial risk of infection. However, a few limitations exist in this original research. Firstly, due to poor data management system in our part of the world, we were unable to determine the cholera trends in healthcare facilities in Enugu metropolis. Secondly, although our study confirmed culturable cholera hazard via PCR and calculated the probability risk of infection using the β-Poisson dose response models, no animal studies were carried out to determine if the gene product of ctx in the strains detected in this study is capable of engendering illness (cholera). Furthermore, the data presented in this study may represent an underestimate of Vibrio abundance in the Asata River due to the use of culturable V. cholerae isolation techniques. It is worth noting that other viable but non-culturable (VBNC) strains may also play an important role in disease transmission.
Again, the QMRA for exposure through drinking water was carried out with a conservative volume 1000 ml of untreated river water consumed. The estimated risks in this study may well be lower than the actual, knowing that WHO recommended a daily consumption of 2000 ml.
Data availability
All data generated and analysed during this current study are included in this published article.
References
UNICEF/WHO. Progress on household drinking water, sanitation and hygiene, 2000–2017. 2019. https://washdata.org/sites/default/files/documents/reports/2019-07/jmp-2019-wash-households.pdf. Accessed 7 Mar 2022
United Nations. United Nations Sustainable Development Goal 6: Ensure access to water and sanitation for all. 2022. https://www.un.org/sustainabledevelopment/water-and-sanitation/https://www.un.org/sustainabledevelopment/water-and-sanitation/. Accessed 5 Mar 2022.
Ramirez-Castillo FY, Loera-Muro A, Jacques M, Garneau P, Avelar-Gonzalez FJ, Harel J, Guerrero-Barrera AL. Waterborne pathogens: detection methods and challenges. Pathogens. 2015;4(2):307–34. https://doi.org/10.3390/pathogens4020307.
Huq A, Haley BJ, Taviani E, Chen A, Hasan NA, Cowell RR. Detection, isolation, and identification of Vibrio cholerae from the environment. Curr Protoc Microbiol. 2013. https://doi.org/10.1002/9780471729259.mc06a05s26.Detection.
GBD. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17(9):909–48. https://doi.org/10.1016/S1473-3099(17)30276-1.
Ali M, Nelson AR, Lopez AL, Sack DA. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis. 2015;9(6):1–13. https://doi.org/10.1371/journal.pntd.0003832.
Deshayes S, Daurel C, Cattoir V, Parienti JJ, Quilici ML, de La Blanchardière A. Non-O1, non-O139 Vibrio cholerae bacteraemia: case report and literature review. Springerplus. 2015;4(1):575. https://doi.org/10.1186/s40064-015-1346-3.
Harris JB, Khan AI, LaRocque RC, Dorer DJ, Chowdhury F, Faruque ASG, Sack DA, Ryan ET, Qadri F, Calderwood SB. Blood group, immunity, and risk of infection with Vibrio cholerae in an area of endemicity. Infect Immun. 2005;73(11):7422–7. https://doi.org/10.1128/IAI.73.11.7422-7427.2005.
Adagbada AO, Adesida SA, Nwaokorie FO, Niemogha MT, Coker AO. Cholera epidemiology in Nigeria: an overview. Pan Afr Med J. 2012. https://doi.org/10.11604/pamj.2012.12.59.1627.
Legros D. Global cholera epidemiology: opportunities to reduce the burden of cholera by 2030. J Infect Dis. 2018;218(3):S137–40. https://doi.org/10.1093/infdis/jiy486.
GTFCC. Global Task Force on Cholera Control. Technical Note Use of antibiotics for the treatment and control of cholera. 2018. https://www.gtfcc.org/wp-content/uploads/2019/10/gtfcc-technical-note-on-use-of-antibiotics-for-the-treatment-of-cholera.pdf. Accessed 2 Apr 2022.
Weber JT, Mintz ED, Greene KD, Puhr ND, Cameron DN, Barrett TJ, Tauxe RV, Blake PA, Canizres R, Semiglia A, Gomez I, Sempertegui R, Davila A, Tenover FC, Bean NH, Ivey C. Epidemic cholera in Ecuador: multidrug–resistance and transmission by water and seafood. Epidemiol Infect. 1994;112(1):1–11. https://doi.org/10.1017/S0950268800057368.
WHO/WER. Cholera 2016: WHO|Weekly Epidemiological Record. 2017;92(35):501–20. WHO. 2017. https://www.who.int/wer/2017/wer9235/en/. Accessed 12 Mar 2022.
NCDC. Nigeria Centre for Disease Control (NCDC): Weekly Epidemiological Report: Preparing to respond to a Cholera Outbreak. 2017;7:2. https://reliefweb.int/report/nigeria/ncdc-weekly-epidemiological-report-volume-7-no-22-16-june-2017. Accessed 12 June 2022.
Ajibade I, McBean G, Bezner-Kerr R. Urban flooding in Lagos, Nigeria: patterns of vulnerability and resilience among women. Glob Environ Chang. 2013;23(6):1714–25.
Abdussalam AF. A spatial autocorrelation analysis of cholera patterns and social risk factors in Nigeria. Acad Res Int. 2015;6(4):136–47.
NCDC. Nigeria Centre for Disease Control (NCDC): Cholera Situation Report Weekly Epidemiological Report 29. 2022. https://reliefweb.int/report/nigeria/ncdc-cholera-situation-report-weekly-epidemiological-report-29-epi-week-52-27. Accessed 10 July 2022.
Elimian K, Yennan S, Musah A, Cheshi LD, King C, Dunkwu L, Mohammed AL, Ekeng E, Akande OW, Ayres S, Gandi B, Pembi E, Saleh F, Omar AN, Crawford E, Olopha OO, Nnaji R, Muhammad B, Luka-Lawal R, Ihueze AC, Olatunji D, Ojukwu C, Akinpelu AM, Adaga E, Abubakar Y, Nwadiuto I, Ngishe S, Alowooye AB, Nwogwugwu PC, Kamaldeen K, Abah HN, Chukwuebuka EH, Yusuff HA, Mamadu I, Mohammed AA, Peter S, Abbah OC, Oladotun PM, Oifoh S, Olugbile M, Agogo E, Ndodo N, Babatunde O, Mba N, Oladejo J, Ilori E, Alfven T, Myles P, Ochu CL, Ihekweazu C, Adetifa I. Epidemiology, diagnostics and factors associated with mortality during a cholera epidemic in Nigeria, October 2020–October 2021: a retrospective analysis of national surveillance data. BMJ Open. 2022;12(9): e063703.
Alam MT, Weppelmann TA, Weber CD, Johnson JA, Rashid MH, Birch CS, Brumback BA, De Rochars VEMB, Morris GJ, Ali A. Monitoring water sources for environmental reservoirs of toxigenic Vibrio cholerae O1, Haiti. Emerg Infect Dis. 2014;20(3):356–63. https://doi.org/10.3201/eid2003.131293.
Pang B, Yan M, Cui Z, Ye X, Diao B, Ren Y, Gao LZ, Kan B. Genetic diversity of toxigenic and nontoxigenic Vibrio cholerae serogroups O1 and O139 revealed by array-based comparative genomic hybridization. J Bacteriol. 2007;189(13):4837–49. https://doi.org/10.1128/JB.01959-06.
Cabral JPS. Water microbiology: bacterial pathogens and water. Int J Environ Res Public Health. 2010;7:3657–703. https://doi.org/10.3390/ijerph7103657.
Gennari M, Ghidini V, Caburlotto G, Lleo MM. Virulence genes and pathogenicity islands in environmental Vibrio strains nonpathogenic to humans. FEMS Microbiol Ecol. 2012;82(3):563–73. https://doi.org/10.1111/j.1574-6941.2012.01427.x.
Kolawole O, Ibitayo A, Mustapha S, Olubayode A. The incidence of Vibrio cholerae as an indicator of pollution of Oyun River in Ilorin, Kwara State Nigeria. Egypt Acad J Biol Sci. 2020;12(1):45–54. https://doi.org/10.21608/eajbsg.2020.82018.
Bulus GH, Ado SA, Yakubu SE, Ella EE. Isolation and characterization of Vibrio cholerae from water sources in Zaria, Nigeria. Ann Exp Biol. 2015;3(3):8–13.
Chikwendu C. Multiple in antimicrobial resistance Vibrio spp isolated from river and aquaculture water sources in Imo State, Nigeria. Br Microbiol Res J. 2014;4(5):560–9. https://doi.org/10.9734/bmrj/2014/4896.
Oyedeji KS, Niemogha MT, Nwaokorie FO, Bamidele TA, Ochoga M, Akinsinde KA, Brai BI, Oladele D, Omonigbehin EA, Bamidele M, Fesobi TW, Musa AZ, Adeneye AK, Smith SI, Ujah IA. Molecular characterization of the circulating strains of Vibrio cholerae during 2010 cholera outbreak in Nigeria. J Health Popul Nutr. 2013;31(2):178–84. https://doi.org/10.3329/jhpn.v31i2.16381.
Okoh BE. Antibiotic susceptibility patterns and plasmid profile of Vibrio cholerae from water samples in Elele Community, Rivers State, Nigeria. J Appl Sci Environ Manag. 2012;16(1):115–20.
Eja ME, Abriba C, Etok CA, Ikpeme EM, Arikpo GE, Enyi-Idoh KH, Ofor UA. Seasonal occurrence of Vibrios in water and shellfish obtained from the Great Kwa River estuary, Calabar, Nigeria. Bull Environ Contam Toxicol. 2008;81(3):245–8. https://doi.org/10.1007/s00128-008-9482-x.
Shittu OB, Olaitan JO, Amusa TS. Physico-chemical and bacteriological analyses of water used for drinking and swimming purposes in Abeokuta, Nigeria. Afr J Biomed Res. 2008;11(3):285–90. https://doi.org/10.4314/ajbr.v11i3.50739.
Idika IN, Audu R, Oyedeji K, Iyanda R, Egbom CA. Investigation of different water sources as a possible cause of cholera outbreak in Lagos in 1997. J Niger Infect Control Assoc. 2000;3(2):6–8. https://doi.org/10.4314/jnica.v3i2.10717.
Ngwa MC, Wondimagegnehu A, Okudo I, Owili C, Ugochukwu U, Clement P, Devaux I, Pezzoli L, Ihekweazu C, Jimme MA, Winch P, Sack DA. The multi-sectorial emergency response to a cholera outbreak in Internally Displaced Persons camps in Borno State, Nigeria, 2017. BMJ Glob Health. 2020;5(1):1–12. https://doi.org/10.1136/bmjgh-2019-002000.
Dan-Nwafor CC, Ogbonna U, Onyiah P, Gidado S, Adebobola B, Nguku P, Nsubuga P. A cholera outbreak in a rural north central Nigerian community: an unmatched case-control study. BMC Public Health. 2019;19(1):112–8. https://doi.org/10.1186/s12889-018-6299-3.
Osinowo OO. Water quality assessment of the Asata River catchment area in Enugu Metropolis, Southeast Nigeria. J Afr Earth Sc. 2016;2016(121):247–54. https://doi.org/10.1016/j.jafrearsci.2016.06.009.
Ozochi CA, Nwankwo CEI, Enemuor SC, Chidebelu PE, Adukwu EC, Chigor VN. Variations in bacteriological and physicochemical water quality characteristics of Asata River, Enugu Nigeria. Microbiol J. 2023;13:1–10.
Kokashvili T, Whitehouse CA, Tskhvediani A, Grim CJ, Elbakidze T, Mitaishvili N, Janelidze N, Jaiani E, Haley BJ, Lashkhi N, Huq A, Colwell RR, Tediashvili M. Occurrence and diversity of clinically important Vibrio species in the aquatic environment of Georgia. Front Public Health. 2015;3(Oct):1–12. https://doi.org/10.3389/fpubh.2015.00232.
Nongogo V, Okoh AI. Occurrence of Vibrio pathotypes in the final effluents of five wastewater treatment plants in Amathole and Chris Hani District municipalities in South Africa. Int J Environ Res Public Health. 2014;11(8):7755–66. https://doi.org/10.3390/ijerph110807755.
Dickinson G, Lim K-Y, Jiang SC. Quantitative microbial risk assessment of pathogenic Vibrios in marine recreational waters of Southern California. Appl Environ Microbiol. 2013;79(1):294–302. https://doi.org/10.1128/AEM.02674-12.
Chun J, Huq A, Colwell RR. Analysis of 16S–23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl Environ Microbiol. 1999;65(5):2202–8. https://doi.org/10.1128/aem.65.5.2202-2208.1999.
Fields PI, Popovic T, Wachsmuth K, Olsvik O. Use of polymerase chain reaction for detection of toxigenic Vibrio cholerae O1 strains from the Latin American cholera epidemic. J Clin Microbiol. 1992;30(8):2118–21. https://doi.org/10.1128/jcm.30.8.2118-2121.1992.
Brouwer AF, Masters NB, Eisenberg JNS. Quantitative microbial risk assessment and infectious disease transmission modeling of waterborne enteric pathogens. Curr Environ Health Rep. 2018;5:293–304. https://doi.org/10.1007/s40572-018-0196-x.
Chigor VN, Sibanda T, Okoh AI. Assessment of human health risks from adenoviruses, hepatitis A virus, rotaviruses and enteroviruses in the Buffalo River and three source water dams in the Eastern Cape Province, South Africa. Food Environ Virol. 2014;6:87–98. https://doi.org/10.1007/s12560-014-9138-4.
Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, Martinez-Urtaza J. Vibrio spp. infections. Nat Rev Dis Primers. 2018. https://doi.org/10.1038/s41572-018-0005-8.
West PA. The human pathogenic vibrios—a public health update with environmental perspectives. Epidemiol Infect. 1989;103(1):1–34.
WHO. Quantitative microbial risk assessment: application for water safety management. WHO Document Production Services, Geneva, Switzerland. 2016; https://www.who.int/publications/i/item/9789241565370. Accessed 20 Mar 2020.
Hass CN, Rose JB, Gerba CP. Quantitative microbial health risk assessment. New York: John Willy and Sons Inc; 1999.
U.S. Environmental Protection Agency and U.S. Department of Agriculture/Food Safety and Inspection Service (USEPA&USDA/FSIS). Microbial risk assessment guideline: pathogenic microorganisms with focus on food and water. EPA/100/J-12/001; USDA/FSIS/2012-001. 2012.
Brumfield KD, Chen AJ, Gangwar M, Usmani M, Hasan NA, Jutla AS, Huq A, Colwell RR. Environmental factors influencing occurrence of Vibrio parahaemolyticus and Vibrio vulnificus. Appl Environ Microbiol. 2023;89(6):1–19. https://doi.org/10.1128/aem.00307-23.
Pande G, Kwesiga B, Bwire G, Kalyebi P, Riolexus AA, Matovu JKB, Makumbi F, Mugerwa S, Musinguzi J, Wanyenze RK, Zhu BP. Cholera outbreak caused by drinking contaminated water from a lakeshore water-collection site, Kasese District, south-western Uganda, June–July 2015. PLoS ONE. 2018;13(6): e0198431. https://doi.org/10.1371/journal.pone.0198431.
Odjadjare EEO, Igbinosa EO. Multidrug-resistaFnt Vibrio species isolated from abattoir effluents in Nigeria. J Infect Dev Ctries. 2017;11(5):373–8. https://doi.org/10.3855/jidc.8097.
Igbinosa EO, Obi LC, Tom M, Okoh AI. Detection of potential risk of wastewater effluents for transmission of antibiotic resistance from Vibrio species as a reservoir in a peri-urban community in South Africa. Int J Environ Health Res. 2011;21(6):402–14. https://doi.org/10.1080/09603123.2011.572278.
Kirschner AKT, Pleininger S, Jakwerth S, Rehak S, Farnleitner AH, Huhulescu S, Indra A. Application of three different methods to determine the prevalence, the abundance and the environmental drivers of culturable Vibrio cholerae in fresh and brackish bathing waters. J Appl Microbiol. 2018;125(4):1186–98. https://doi.org/10.1111/jam.13940.
Ramamurthy T, Chowdhury G, Pazhani GP, Shinoda S. Vibrio fluvialis: an emerging human pathogen. Front Microbiol. 2014;5(91):1–8. https://doi.org/10.3389/fmicb.2014.00091.
Funding
No funding was received for this study.
Author information
Authors and Affiliations
Contributions
Conceptualization: CVN, OCA. Investigation: OCA, OCC. Data curation and data analysis: OCA, OCC, CVN. Original draft preparation: OCA. Supervision: CVN, AEC, UVC. Writing, Reviewing and Editing: OCA, EMC, OCC, UVC, CVN. All authors read and approved the final draft.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ozochi, C.A., Okonkwo, C.C., Adukwu, E.C. et al. Detection and quantitative microbial risk assessment of pathogenic Vibrio cholerae in a river used for drinking, domestic, fresh produce irrigation and recreational purposes. Discov Water 4, 7 (2024). https://doi.org/10.1007/s43832-024-00059-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43832-024-00059-z