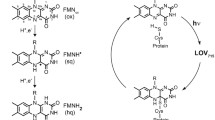
Abstract
Light, oxygen, voltage (LOV) photoreceptors are widely distributed throughout all kingdoms of life, and have in recent years, due to their modular nature, been broadly used as sensor domains for the construction of optogenetic tools. For understanding photoreceptor function as well as for optogenetic tool design and fine-tuning, a detailed knowledge of the photophysics, photochemistry, and structural changes underlying the LOV signaling paradigm is instrumental. Mutations that alter the lifetime of the photo-adduct signaling state represent a convenient handle to tune LOV sensor on/off kinetics and, thus, steady-state on/off equilibria of the photoreceptor (or optogenetic switch). Such mutations, however, should ideally only influence sensor kinetics, while being benign with regard to the nature of the structural changes that are induced by illumination, i.e., they should not result in a disruption of signal transduction. In the present study, we identify a conserved hydrophobic pocket for which mutations have a strong impact on the adduct-state lifetime across different LOV photoreceptor families. Using the slow cycling bacterial short LOV photoreceptor PpSB1-LOV, we show that the I48T mutation within this pocket, which accelerates adduct rupture, is otherwise structurally and mechanistically benign, i.e., light-induced structural changes, as probed by NMR spectroscopy and X-ray crystallography, are not altered in the variant. Additional mutations within the pocket of PpSB1-LOV and the introduction of homologous mutations in the LOV photoreceptor YtvA of Bacillus subtilis and the Avena sativa LOV2 domain result in similarly altered kinetics. Given the conserved nature of the corresponding structural region, the here identified mutations should find application in dark-recovery tuning of optogenetic tools and LOV photoreceptors, alike.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Almost all organisms possess photoreceptor proteins that enable them to perceive and respond to light as a physical stimulus. At present, at least ten photoreceptor protein families can be distinguished [1,2,3,4,5,6,7,8,9]. Each of them reacts to light of a defined wavelength range utilizing a characteristic photochemistry to drive protein structural changes, which in turn trigger a specific physiological response. One family, widely distributed throughout the three kingdoms of life (with the exception of Animalia) are so-called light, oxygen, voltage (LOV) photoreceptor proteins [10], which sense UV/blue light via small, structurally conserved protein domains, called LOV domains [11, 12]. LOV domains belong to the Per-ARNT-Sim (PAS) family and bind flavins (flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD) or rarely riboflavin (RF)) as blue light-absorbing chromophores [13, 14]. Plant LOV photoreceptors mediate physiological effects such as phototropism, light-induced stomatal opening, light-controlled chloroplast movement, leaf photomorphogenesis and circadian responses [15]. In green algae such as Chlamydomonas reinhardtii, sexual differentiation, phototaxis, photoprotection, gene expression and eyespot size are regulated by a phototropin LOV photoreceptor [16,17,18,19,20], while in stramenopile algae such as Vaucheria frigida, the LOV photoreceptor aureochrome is responsible for controlling photomorphogenesis [21]. In fungi, photoadaptation, i.e., carotenoid biosynthesis, is controlled by the LOV photoreceptors VVD and WC-1 [22]. In prokaryotes, functions are equally diverse, including control of the general stress response machinery, virulence, cell–cell adhesion, the synthesis and degradation of secondary messengers and the formation of photosynthesis pigments [6, 23, 24]. Interestingly, all LOV photoreceptor signaling responses, as diverse as they may be, hinge on the same photophysical and photochemical principles that constitute the so-called LOV photocycle. Light absorption by the LOV domain’s flavin chromophore, which is non-covalently bound within the LOV domain in the dark, induces a series of photophysical and photochemical reactions that eventually lead to the formation of a covalent adduct between a totally conserved cysteine residue of the protein and the flavin chromophore, accompanied by protonation of the FMN-N5 atom. This localized structural change induces large-scale structural changes throughout the LOV sensory domain, to N and/or C-terminally fused effector domains that control different signaling cascades and, thus, enable a large number of different physiological reactions [2]. In addition, LOV photoreceptors lacking fused effector domains can also be identified. Best characterized examples include the two short LOV proteins PpSB1-LOV and PpSB2-LOV from Pseudomonas putida [25,26,27,28,29], the short LOV protein DsLOV from Dinoroseobacter shibae [23], the short LOV photoreceptor RsLOV of Rhodobacter sphaeroides [30] and the short LOV protein VVD from Neurospora crassa [31]. Interestingly, all of the so far-identified short LOV proteins are suggested to be involved in the regulation of gene expression [23, 32, 33] likely utilizing short N- and/or C-terminal α-helical extensions to regulate LOV–LOV dimerization or alterations in the mode of dimerization in a light-dependent manner [23, 28, 34], which in turn might influence the interaction with downstream effectors. This architectural plasticity, manifested in the modular nature of LOV photoreceptors, has led in recent years to the widespread use of LOV domains as optogenetic tools, i.e., for monitoring and perturbation of biological functions with high spatiotemporal resolution [35]. For efficient photoswitching and, thus, also for the development of optogenetic switches, not only the structural changes trigged by illumination are of importance, but also the kinetics of the different photocycle steps, e.g., those that determine the stability of the signaling-active conformation, which can either be the light or dark state [23, 35,36,37,38,39,40]. Therefore, a detailed mechanistic understanding of both LOV photocycle kinetics and the underlying light-dependent structural changes of the photoreceptor is essential for understanding photoreceptor function and the development of optogenetic tools. While the above-mentioned adduct-formation step happens in most LOV domains in the time range of ∼200 ns–2 μs [41,42,43,44,45], the recovery of the initial dark state, which involves thermal breaking of the flavin-cysteinyl covalent linkage and deprotonation of the FMN-N5 atom, can take from seconds to days depending on the LOV protein [12, 23, 25, 46,47,48,49,50]. The latter step is therefore limiting for efficient photoswitching, as it determines the steady-state on/off equilibria of the photoreceptor (or optogenetic switch) at a given light intensity and illumination time [37]. For optogenetic tool development and optimization, it is, therefore, important, that any variants, which show altered kinetics, are still undergoing the same light-dependent structural changes as the native photoreceptor. Probing such an impact [51,52,53,54], however, has been neglected in many earlier studies, not the least due to the lack of a simple functional or biophysical read-out [52, 55]. To date, several factors have been described that tune the dark recovery of LOV photoreceptors. Those include (i) alteration of the H-bonding network at the pyrimidine side of the flavin ring system [56, 57], (ii) steric factors that impact the conformational freedom of the adduct-forming cysteine residue [58, 59] or of the FMN molecule itself [47, 60] and iii) factors that influence FMN-N5 deprotonation such as solvent access or base catalysis [57, 59, 61,62,63]. Despite these efforts, the structural and mechanistic basis of LOV domain dark-recovery kinetic variability has still not been fully understood. Moreover, the transferability of mutations that tune the dark recovery has only been tested in a few cases in different LOV photoreceptors [51, 59, 64] and the impact of the corresponding mutations on the light-induced structural changes, and thus photoreceptor functionality, are still insufficiently characterized.
To address these issues, we chose the slow cycling LOV protein PpSB1-LOV [25], which is a well-characterized model system for bacterial short LOV proteins [27,28,29]. Based on previous work [28], we here identified one residue, I48, localized in a conserved hydrophobic pocket, which when mutated to threonine resulted in a three times faster dark-recovery reaction as compared to the wild-type protein, while retaining wild-type like flavin-loading and light-induced structural changes, as probed here by NMR spectroscopy and X-ray crystallography. Introduction of the corresponding mutation in Bacillus subtilis YtvA (I57T) and the Avena sativa phototropin-1 LOV2 domain (AsLOV2) (I445T) also resulted in altered dark-recovery kinetics, while the introduction of further mutations within the hydrophobic I48 pocket led to variants with up to 32-fold faster dark recovery. Our study, thus, stresses the general importance of the corresponding structural region for tuning of the dark recovery of LOV photoreceptors, while at the same time opening up new possibilities for the fine-tuning of LOV-based optogenetic tools.
2 Materials and methods
2.1 Bacterial strains and plasmids
All bacterial strains and plasmids that were used in this study are listed in Table S1. Escherichia coli DH5 and E. coli BL21(DE3) were used for cloning purposes and heterologous overexpression, respectively.
2.2 Cloning and site-directed mutagenesis
All oligonucleotides used for site-directed mutagenesis are given in Table S2. The variants PpSB1-LOV-I48T, -F55H, -F55A, -V33T, -Y43A and -Y43H as well as the YtvA-I57T variant (equivalent to I48T and I445T in PpSB1-LOV and AsLOV2, respectively) were generated by Quikchange PCR according to the instructions given by the manufacturer (Stratagene, La Jolla, CA) using pET28a-PpSB1-LOV [25] and pET28a-YtvA [65] as templates, respectively. The gene coding for the LOV2 domain of the Avena sativa phototropin-1 (AsLOV2; residues 404–546 of full-length phototropin-1) as well as the corresponding gene coding for the AsLOV-I445T variant were obtained by gene synthesis (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Both synthetic genes, appended with a 5’ NdeI and a 3’ SalI restriction endonuclease recognition site, were subcloned from the respective pMA-RQ(+) synthesis vectors (Table S1) into pET28a using NdeI and SalI. All constructs were appended with an N-terminal hexa-histidine tag (sequence: MGSSHHHHHHSSGLVPRGSH) encoded by the expression vector (Table S1). All final constructs were verified by sequencing (SeqLab, Göttingen, Germany).
2.3 Heterologous gene expression and protein purification
Expression and purification of wild-type PpSB1-LOV and its variants was carried out with minor modifications as described previously [28]. All samples were either prepared without N-terminal hexa-histidine tag; removing the tag by proteolytic cleavage with thrombin [28] (NMR studies), or were used as His-tagged variants (UV/Vis spectroscopic characterization of all PpSB1-LOV variants, YtvA, YtvA-I47T, AsLOV2 and AsLOV2-I445T, crystallization of PpSB1-LOV-I48T). Protein samples were desalted using PD-10 desalting columns (size: 1.4 × 5 cm, gel bed volume: 8.3 mL; GE-Healthcare, Chicago, Illinois, USA) and transferred to 20 m M sodium phosphate buffer pH 6.4 supplemented with 10 mM NaCl and 1 mM DTT as storage buffer. Samples were stored either in soluble form at 4 °C or as lyophilized powder at – 20 °C in the dark until further use.
2.4 UV–vis spectroscopy
UV/Vis spectrophotometric measurements were carried out under dim-red safety light using a Cary-60 UV/Vis spectrophotometer (Agilent Technologies, Santa Clara, CA, USA) equipped with a single-cell cuvette holder thermostatted to 40 ± 2 °C (PpSB1-LOV and its variants) or 25 ± 1 °C (YtvA, YtvA-I57T, AsLOV2, AsLOV2-I445T). All measurements were performed in 20 mM sodium phosphate buffer (pH 6.4) supplemented with 10 mM NaCl, 1 mM DTT, 0.03% (w/v) NaN3 in a 90%/10% (vol/vol) H2O/D2O mixture, to mimic the conditions during the NMR measurement. All samples were diluted to an absorbance of 0.1 at λmaxFMN = 450 nm. Samples were illuminated using a blue light-emitting high-power LED (Luxeon Lumileds, Phillips, Aachen, Germany; 2.6 mW/cm2) mounted on top of the cuvette. Illumination was controlled using an Arduino UNO (Smart Projects, Italy) microcontroller as described previously [52, 66]. After illumination, dark-state recovery was either followed by recording sequential UV/Vis spectra (PpSB1-LOV, YtvA and their variants) or by directly following the recovery of the absorbance at 485 nm (AsLOV2 and AsLOV2-I445T). The adduct-state life time τRec was obtained by plotting the recovery of the absorbance at 485 nm against the time and fitting the data to a mono-exponential decay function according to:
where y is the absorbance at 485 nm, y(t = 0) is the starting absorbance at t = 0 and τrec is the adduct-state lifetime.
2.5 NMR spectroscopy
[U-13C,15 N] and [U-15 N] PpSB1-LOV wild type and the I48T variant samples were prepared in 20 mM sodium phosphate buffer (pH 6.4) supplemented with 10 mM NaCl, 1 mm DTT, 0.03% (w/v) NaN3 in a 90%/10% (vol/vol) H2O/D2O mixture. The protein concentration was 500 µM unless stated otherwise. NMR spectra were recorded at 40 °C on spectrometers operating at protein frequencies of 600, 700, and 800 MHz, equipped with cryogenically cooled z-gradient probes. The sample temperature was calibrated using a perdeuterated methanol sample as described by Findeisen et al. [67]. For measurement of light-state samples, the NMR sample transferred inside a 5 mm NMR Shigemi tube was pre-illuminated with blue light using a high-power LED (Luxeon Lumileds, Royal Blue λmax = 450 nm, 2.6 mW cm−2; Philips GmbH, Aachen, Germany). The sample was kept illuminated for the course of experiments with a fiber-optic cable, placed inside the Shigemi plunger, which transmits the blue light through a collimator lens. This mode of illumination inevitably results in lower light intensity during actual the NMR experiment. For PpSB1-LOV wild type, this mode of illumination was sufficient to hold the whole protein sample in the light state to enable triple resonances experiments to acquire assignments [28]. The Shigemi tube with the attached fiber-optic cable was then lowered inside the magnet.
2.5.1 Chemical shift perturbation analysis
PpSB1-LOV wild-type backbone resonance assignments were determined previously [28]. The backbone assignments for the dark state of the PpSB1-LOV- I48T variant were confirmed using a 3D HNCA experiment [68]. Spectra were processed using the NMRpipe package [69] and analyzed with CcpNMR analysis [70]. The weighted average chemical shift differences, (Δδav), were calculated using the formula: Δδav = [(Δδ2HN + Δδ2N/25)/2]1/2 [71].
2.6 Crystallization of PpSB1-LOV-I48T, data collection, and analysis
2.6.1 Protein crystallization
The purified PpSB1-LOV-I48T protein in 20 mM sodium phosphate buffer at pH 6.4 supplemented with 10 mM NaCl was concentrated to 15 mg/mL using 10 kDA molecular weight cut-off centrifugal concentrators (Macrosep Advance, Pall, Port Washington, NY, USA). Two parallel crystallization setups were performed at 19 °C—One in complete darkness, and the other under continuous illumination with blue-light LED arrays (λmax = 450 nm, Luxeon Lumileds, Phillips, Aachen, Germany). Both approaches were done using the sitting-drop vapor diffusion method using 1 μL of protein and 1 μL of the reservoir solution. The protein crystallized in the dark under the conditions 0.2 M sodium chloride, 0.1 M MES pH 6.0, 15% (v/v) pentaerythritol propoxylate in the monoclinic symmetry, while under constant light conditions the crystals appeared in hexagonal symmetry under the conditions 0.085 M HEPES pH 7.5, 17% (w/v) PEG 4000, 15% (v/v) glycerol, 8.5% (v/v) isopropanol. The respective crystallization time was 2–3 weeks in both cases.
2.6.2 Data collection and structure determination
Both the crystal types were cryo-compatible in their respective reservoir solutions and were used as such for data collection at 100 K. Diffraction data were recorded at beamlines ID23-1 and ID29 at ESRF (Grenoble, France, [72, 73]). The respective wavelengths and the corresponding detector types of each beamline can be found in Table 1. Data collection strategies were established using the program BEST [74] such that the radiation damage was minimized while ensuring maximum data completeness. Data processing was carried out using the program XDS [75] and AIMLESS (part of the CCP 4 package, [76]). The initial phases were obtained by molecular replacement using MOLREP (CCP4-package., [76]), and the respective models were generated from the crystal structures of PpSB1-LOV in the dark state (PDB-ID: 5J3W) and PpSB1-LOV in the light-state (PDB-ID: 3SW1). Four molecules occupy the asymmetric unit in the monoclinic crystal form of the dark state with space group C2, while the blue light-excited crystal structure with space group P6122 has one molecule in the asymmetric unit. The respective Matthews coefficients [77] are 2.2 and 2.55 Å3/Da, corresponding to a solvent content of 44.2% and 51.8%. Both X-ray structures were further refined with the program Phenix [61] and visually inspected with the graphics program Coot [78] and corrected accordingly. The statistics are shown in detail in Supporting Table S3.
2.7 Sequence conservation analysis
The conservation of residues constituting the hydrophobic I48 pocket was analyzed as described previously [52, 79] using the comprehensive LOV domain sequence data set provided by Glantz and co-workers [80]. Sequence logos were generated with WebLogo 3 [81] and analyzed with Skylign [82]. In the latter case, conservation was quantified as observed counts relying on sequence weights to account for a high degree of similarity in a subset of sequences. This yields for each amino acid position probabilities for the occurrence of the possible different amino acids, reflecting the overall sequence diversity of the data set.
3 Results
Our previous NMR analysis to investigate the light-induced conformational changes in the PpSB1-LOV wild-type protein revealed that residue I48, located at the C-terminal end of the Dα helix, shows a remarkable chemical shift perturbation upon illumination with blue light [28]. Such a large change, indicative of a major conformational and/or environmental change around I48, is surprising since this residue is located at a distance of ~ 10 Å away from the FMN chromophore and the dimer interface, with the latter experiencing the most pronounced chemical shift perturbation [28], structurally manifested in a rotation of the two dimer subunits relative to each other, as observed in the dark- and light-state crystal structures of PpSB1-LOV [28].
As judged from the corresponding dark and light-state structures, illumination with blue light leads to a rotation of ~ 120° around the χ1 angle of I48 (Fig. 1 B). A closer inspection reveals that the region around the I48-haboring Dα helix is primarily composed of hydrophobic residues located on Bβ, Cα, Eα, and Dα. For simplicity, we will refer in the following to this structural feature as the “I48 pocket” (Fig. 1B). This residue corresponds to I57 in B. subtilis YtvA and I445 in Avena sativa AsLOV2. For simplicity, we will in the following refer to this pocket as the I48 pocket. The I48 pocket hereby stabilizes the Eα helix and the adduct-forming cysteine (C53 in PpSB1-LOV) located within this helix. The pocket is dominated by residues L30 (I39 in YtvA, I427 in AsLOV2), V33 (V42 in YtvA, A430 in AsLOV2), F37 (F46 in YtvA, F434 in AsLOV2), Y43 (Y52 in YtvA, Y440 in AsLOV), I48 (I57 in YtvA, I445 in AsLOV2), F55 (F64 in YtvA, F452 in AsLOV2, and L56 (L65 in YtvA, L453 in AsLOV2), in both light and dark states of the protein (Fig. 1B). Amongst these residues, the side chains of V33, Y43, I48, and F55 are at a distance of more than 6 Å from the FMN chromophore.
A Crystal structure of the PpSB1-LOV dimer in the dark state (PDB: 5J3W) with FMN in stick representation, with carbon atoms in gray, oxygen in red, nitrogen in blue and phosphorus in orange. In addition, I48 is shown as stick representation with carbon atoms in gray. B A close-up of the hydrophobic pocket is depicted as inset. The view shows both the dark- and light-state structure highlighted as transparent cartoon and marks the structural changes of I48 due to illumination. I48 is shown in its dark- and light-state conformation with carbon atoms in dark gray and dark blue, respectively. Residues that constitute the hydrophobic pocket around I48 are shown in their dark-state conformation in stick representation
3.1 The I48T mutation has no impact on the light-induced conformational changes of PpSB1-LOV in solution
To probe the importance of the I48 pocket for the function of PpSB1-LOV, we introduced a hydrophilic residue, threonine, at position 48 of PpSB1-LOV, resulting in the variant PpSB1-LOV-I48T. Threonine was chosen to perturb the I48 pocket, as a result of the hydrophilic side chain γ1OH group and a 45% lower van-der-Waals volume in comparison to isoleucine. The PpSB1-LOV-I48T variant could readily be purified (FMN load > 99% FMN), as estimated from the dark-state UV/Vis spectrum of the protein (Fig. 2A) and reverse-phase UPLC analyses (Fig. 2B). Moreover, wild-type like photocycling is retained in the variant (Fig. 2A), indicative of a fully functional photoreceptor.
A Dark (black line) and light-state (blue line) UV/Vis spectra of PpSB1-LOV-I48T and B UPLC analysis of flavin-species distribution of purified PpSB1-LOV-I48T. The employed UPLC method is described in the Supporting Materials and Methods section. (B, upper panel) UPLC chromatogram of a mixture of authentic, flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), riboflavin (RF) and lumichrome (LC); used as HPLC standards for identification of protein loaded flavins. (B, lower panel) UPLC chromatogram of the flavin species released from PpSB1-LOV-I48T. Evaluation of the data yielded a flavin-species distribution of 99.5% FMN, 0.1% FAD, 0.1% RF and 0.3% LC. Black dashed lines mark the retention times of FAD, FMN, RF and LC
To investigate whether the I48T mutation leads to large-scale structural perturbation of PpSB1-LOV, thereby impairing light-dependent conformational changes, we compared the 2D 1H-15 N HSQC spectra of wild-type PpSB1-LOV and PpSB1-LOV-I48T (Figure S1 A, B). The backbone 1H and 15 N chemical shift assignments for the dark state of the I48T variant were confirmed using a 3D HNCA experiment.
Confirmation of the light-state chemical shift assignments, however, was not possible for PpSB1-LOV-I48T because the blue-light LED source used for chromophore excitation (see Materials and Methods for details) was not able to hold the I48T variant in the light state for the duration of the 3D HNCA experiment (typically more than one day). Therefore, only the resonances which could be unambiguously assigned from the overlay between 2D 1H-15 N HSQC spectra of wild-type PpSB1-LOV and the I48T variant (Figure S1) were used for chemical shift perturbation analysis. Significantly large chemical shift perturbations (Δδav) above the significance level of (Δδav + 1σ(Δδav); dashed line in Fig. 3) due to the mutation are primarily localized around the mutation site, i.e., the Eα and Dα helices, without any significant effects on other regions of the protein (Fig. 3A, B), suggesting that the mutation does not cause large-scale structural changes in the protein.
Backbone amide chemical shift perturbations of PpSB1-LOV due to I48T mutation in the dark-(A) and light state (B). Chemical shift perturbation Δδav relative to the corresponding dark- and light-state chemical shifts of wild-type PpSB1-LOV. Significant chemical shift changes (Δδav) arising from the I48T mutation are mostly localized around the mutation site (residues 39–51). The significance level (Δδav + 1σ(Δδav) is indicated by the black dashed line. A Δδav of the dark-state spectra with a significance level of 0.08 ppm, and B Δδav of the light-state 1H-15 N HSQC spectra with a significance level of 0.06 ppm. The secondary structure elements are shown on top of the Figure
In conclusion, wild-type like flavin-loading, photochemistry, and NMR data indicate that the I48T mutation is benign with respect to the function of the PpSB1-LOV photoreceptor.
3.2 Dark- and light-state crystal structures of PpSB1-LOV-I48T corroborate the benign nature of the I48T mutation
To gain further insights into the structural consequences of the I48T mutation, we crystallized the I48T variant in the dark and under constant illumination and determined the corresponding dark- and light-state crystal structures. Table S3 summarizes data collection and refinement statistics for all here obtained structures. In the dark, the protein yielded monoclinic crystals in the space group C2, consisting of four molecules in an asymmetric unit. This is very similar to our previously reported dark-state structure of wild-type PpSB1-LOV (backbone root mean-square deviation of 0.37 Å; Fig. 4A) that also crystallized in C2 space group with four molecules in the asymmetric unit forming two dimers, which differed primarily in the orientation of the two protruding Jα helices [28]. Under constant illumination, hexagonal crystals (space group, P 61 2 2) of PpSB1-LOV-I48T were obtained with one molecule per asymmetric unit. However, PISA analysis suggests a wild-type-like dimer arrangement, which agrees with the results from size exclusion chromatography where PpSB1-LOV-I48T elutes as a dimer (Figure S2). Superimposition of the corresponding wild-type light-state structure of PpSB1-LOV yields a backbone RMSD value of 0.37 Å, indicative of nearly identical structures. Thus, the light-dependent conformational changes previously described for PpSB1-LOV wild type still occur in the I48T variant, corroborating the functionally benign nature of the mutation (Fig. 4E). Residue I48 resides at the C-terminal end of Dα helix, forming part of a hydrophobic pocket comprising the residues L29, L30, V33, F37, Y43, L49 and F55 from Bβ, Cα, Eα, and Dα (see inserts to Fig. 4A, B). In PpSB1-LOV I48T crystal structures, these residues show no significant changes when superimposed on the respective wild-type structures (Fig. 4A, B). This is not surprising as they are not directly involved in either FMN coordination or in the formation of dimer interface.
Overlay of dark- and light-state crystal structures of PpSB1-LOV and PpSB1-LOV I48T. A Superposition of dark-state dimer (A–B chains) of PpSB1-LOV I48T (green) and the corresponding dark-state wild-type PpSB1-LOV dimer (B–D chains in PDB-ID 5J3W) (salmon). B Superposition of the wild-type light-state PpSB1-LOV dimer (beige) and the corresponding I48T light-state structure (plum). The insets in panels A and B show the close-up view of the hydrophobic pocket carrying the I48T mutation in dark and light states, respectively. T48 and residues contributing to the hydrophobic pocket that are within 5 Å of I48 are shown as stick models C Superimposition of PpSB1-LOV I48T dark- (green) and light-state structures (plum). For clarity, only the FMN molecule in the respective I48T structure is shown as stick model. In panels A and B, the FMN is colored by element: carbon—yellow; nitrogen—blue; oxygen—red; and phosphorus—magenta
Light-induced changes such as movement of the Dα–Eα loop are similar, where the latter moves out slightly (distance measured between Cα atoms of residue 48 in the two states is ~ 1.6 Å in PpSB1-LOV and 0.9 Å in PpSB1-LOV I48T), and is more solvent exposed in the dark state. We previously reported that the side chain of I48 in the crystal structures of dark and light states of PpSB1-LOV reveals a change in its χ1 angle upon illumination, from χ1 angle of 54° in the dark state to χ1 angle of − 59° in the light state. In contrast, the side chain rotamer of T48 is similar in both, light and dark states in the PpSB1-LOV I48T variant, which is likely due to the shorter side chain. Nevertheless, side chains of both I48 in the wild type and T48 in the corresponding crystal structures point to the hydrophobic pocket, composed of residues mentioned above.
3.3 The hydrophobic I48 pocket is important for dark-recovery tuning
3.3.1 The I48T mutation accelerates the dark recovery of PpSB1-LOV
The inability of the blue-light source to hold the I48T variant in a stable light state during the NMR experiments (see above) indicated that the light state of the I48T variant decays significantly faster than in wild-type PpSB1-LOV. This implies faster adduct rupture and hence a faster dark-recovery process. To quantify this acceleration, we determined the adduct-state lifetime of the I48T variant by UV/Vis spectroscopy by following the change in absorbance at 485 nm. At 40 °C, i.e., to mimic the conditions of our NMR experiments, the I48T variant shows faster dark recovery than wild-type PpSB1-LOV, yielding an adduct-state lifetime of τrec = 25.0 ± 0.8 min, which is about three times faster than the wild type with a τrec = 73.7 ± 0.8 min; under identical conditions (Table 1). Please note that, in previous studies, a τrec of about \(\sim\) 2500 min was determined for PpSB1-LOV at 20 °C [25]. Exemplary UV/Vis time traces can be found in the Supporting Information (Figure S3, A and B; compare X-axis scale).
3.3.2 General importance of the hydrophobic I48 pocket and generality of the mutation for dark-recovery tuning
To infer if the hydrophobic I48 pocket plays a general role in stabilizing the light state, we decided to investigate the effect of mutations of other hydrophobic residues within this pocket on the dark recovery of PpSB1-LOV. We introduced the mutations V33T, Y43A, Y43H and F55A and F55H into PpSB1-LOV. The mutated positions correspond to V42/A430, Y52/Y440 and F64/F452 in YtvA and AsLOV2, respectively. With the exception of the Y43A, which lost its chromophore after purification, and Y43H, which showed a slower dark recovery (τrec = 96.9 ± 1.3 min) compared to the wild type (τrec = 73.7 ± 0.8 min), all tested variants showed a faster dark recovery (Table 1). Exemplary dark-recovery time traces can be found in the Supporting Information (Figure S3, C–F). The most pronounced acceleration was observed for the two F55 variants -F55A (τrec = 2.3 ± 0.1 min) and -F55H (τrec = 3.7 ± 0.1 min), which showed 32- and 20-fold faster dark recoveries, respectively.
Lastly, to test whether the I48T mutation identified to accelerate the dark recovery of PpSB1-LOV, has a similar effect in other LOV photoreceptors, we introduced the I57T and I445T mutations in the YtvA photoreceptor of Bacillus subtilis and the LOV2 domain of the Avena sativa phototropin-1 (AsLOV2), and measured their adduct-state lifetime (Figure S4 and S5) at 25 °C. While the YtvA-I57T variant showed an accelerated dark recovery (τrec = 17.5 ± 0.1 min) compared to wild-type YtvA (τrec = 31.2 ± 8.7 min), surprisingly the corresponding AsLOV2-I445T variant showed a threefold slower dark recovery (τrec = 2.53 ± 0.05 min) as compared to the corresponding wild type (τrec = 0.81 ± 0.02 min).
4 Discussion
Using a combination of UV/Vis spectroscopic methods, NMR spectroscopy and X-ray crystallography, we here identified a residue substitution within a hydrophobic I48 pocket of PpSB1-LOV, which alters the dark recovery of the protein. This observation is in so far surprising, as the mutated residue is located at a distance of about 10 Å from the FMN chromophore and can, thus, only indirectly affect covalent adduct formation between the protein and the flavin chromophore. NMR studies and the here-presented dark- and light-state crystal structures of the faster reverting PpSB1-LOV-I48T variant showed that the introduced mutation is benign with regard to the light-induced conformational changes, and hence most likely does not affect the in vivo functionality of the photoreceptor. Such a feature is important, e.g., for the tuning of LOV-based optogenetic tools, where photocycle kinetics play an important role for the efficacy of the artificially generated light switch [83, 84]. Likewise, LOV photoreceptors with altered photocycle kinetics have recently been used in planta, e.g., for improving biomass production under low light conditions [36]. Based on the additional mutations, which we introduced within the I48 pocket of PpSB1-LOV, we conclude that this structural region is important for dark-recovery tuning in PpSB1-LOV. Moreover, the introduction of similar mutations within two other LOV proteins; the B. subtilis YtvA photoreceptor and the LOV2 domain of A. sativa phototropin-1 (AsLOV2); resulted in variants with altered dark-recovery kinetics. Interestingly, while a faster dark recovery was observed for the YtvA and PpSB1-LOV I57T/I48T variants, respectively, AsLOV2-I445T showed a slower dark recovery. These contradictory trends, although confirming the importance of the hydrophobic I48 pocket for dark-recovery tuning, hint at different tuning mechanisms in AsLOV2 and YtvA/PpSB1-LOV, respectively.
Given the opposite trends seen for the dark recovery of the AsLOV2-I445T, YtvA-I57T and PpSB1-LOV-I48T variants, one would have to assume that placing a threonine into the AsLOV2 pocket stabilizes the FMN-Cys linkage in the light state, while in case of YtvA and PpSB1-LOV a destabilization is observed. However, due to the conserved nature of the hydrophobic I48 pocket (see below), only few structural differences can be identified between PpSB1-LOV, YtvA and AsLOV2 (Figure S4). Superimposing the light-state structures of the isolated LOV domains of YtvA and PpSB1-LOV (both chains) to AsLOV2 yields RMSD values between 1.18 and 2.33 for all backbone atoms (Table S4). While I48 of PpSB1-LOV and I57 of YtvA show very similar light-state conformations, the corresponding I445 of AsLOV2 shows two conformations in the light state, neither matching the conformation seen for YtvA and AsLOV2 (Figure S4), indicative for some flexibility in this region in AsLOV2, which is absent in YtvA and PpSB1-LOV. In addition, both PpSB1-LOV and YtvA possess a valine at position 33 lining the I48 pocket, while AsLOV2 contains a smaller alanine at this position, possibly rendering the pocket more compact in case of PpSB1-LOV and YtvA. Further, in PpSB1-LOV, a glutamine is found at position 38 (on Cα), whose side chain forms an H-bond to backbone of Arg45 (E38-OE1 … R45-N) (on Dα) thereby stabilizing the pocket, while YtvA and AsLOV2 lack such an interaction, as they possess a valine (V47) and leucine (L435) at the corresponding position, respectively. Overall, the hydrophobic pocket might be more compact and less flexible in case of PpSB1-LOV and YtvA compared to AsLOV2. It is tempting to speculate that this feature could account for the opposite behavior of AsLOV2, compared to YtvA and PpSB1-LOV, which was observed for the I445T and I57/T/I48T mutations, respectively.
Mechanistically, introducing a polar threonine side chain into the hydrophobic pocket might impact the dynamics of the overall region, which in turn could impact stability of the FMN-Cys linkage and hence dark recovery. Generation of a L435E/A430V/I445T triple variant of AsLOV2 might help to delineate the nature of observed contradictory trends, while molecular dynamics (MD) simulations of light and dark state of PpSB1-LOV and the corresponding PpSB1-LOV-I48T variant would enable assessment of the dynamics of the corresponding region.
Further supporting the importance of the hydrophobic I48/I57/I445 pocket for dark-recovery tuning, in a previous study the mutation of F46 to histidine in YtvA (corresponding to F37 in PpSB1-LOV and F434 in AsLOV2), was shown to accelerate the dark recovery of the protein 25-fold [6]. Though, when introduced in the artificial photoreceptor YF1, the F46H mutation adversely affected light regulation [51]. When combined with the I39V mutation (corresponding to L30 of the PpSB1-LOV I48 pocket, and I427 of AsLOV2), the recovery of the YtvA F46H/I39V double mutant variant is even accelerated 75-fold [6]. The I427V mutation in AsLOV2 likewise accelerated the dark recovery by a factor of about eight [58]. However, likely due to its close proximity to the FMN chromophore, the YtvA I39V mutation severely impaired light regulation by the YtvA-LOV domain, albeit in the artificial YF1 framework [51]. Those studies corroborate the general importance of the structural region corresponding to the PpSB1-LOV I48 hydrophobic pocket for the dark recovery of LOV proteins in general, while stressing the importance of using distant sites for dark recovery tuning, whose mutation is less likely to influence the signaling response.
4.1 The hydrophobic I48 pocket of PpSB1-LOV is highly conserved
Given the observed mutational effects, the question arises, if the structural region centered around the I48 pocket of PpSB1-LOV is well conserved in other LOV proteins. Based on a multiple-sequence alignment of 1975 LOV domain sequences [79], obtained from the sequence data set of Glantz and co-workers [80], we analyzed the sequence conservation of residues constituting the hydrophobic I48 pocket of PpSB1-LOV by generating a sequence logo (Fig. 5A). For reference, the dark-state structure of PpSB1-LOV is shown in Fig. 5A, B, and an alignment of selected LOV domain sequences is shown in Fig. 5C. In addition, sequence conservation was analyzed with Skylign [82], which for every amino acid position calculates a probability for the occurrence of different amino acids. The corresponding probabilities for the residues constituting the hydrophobic I48 pocket are summarized in Table S5. The pocket is hereby constituted by residues L30 (I39 in YtvA, I427 in AsLOV2), V33 (V42 in YtvA, A430 in AsLOV2), F37 (F46 in YtvA, F434 in AsLOV2), Y43 (Y52 in YtvA, Y440 in AsLOV), I48 (I57 in YtvA, I445 in AsLOV2), F55 (F64 in YtvA, F452 in AsLOV2, and L56 (L65 in YtvA, L453 in AsLOV2) (highlighted with red asterisks in Fig. 5C).
A Sequence logo illustrating the conservation of residues within the hydrophobic I48 pocket. Residues that constitute the pocket are localized on Bβ, Cα, Dα and Eα (red asterisks). Violet asterisks label additional residues that are within 5 Å of I48 but are not part of the I48 pocket of PpSB1-LOV (PDB-ID: 3SW1). The position of I48 of PpSB1-LOV is marked by a red arrow. Residue numbering according to the PpSB1-LOV scheme. The sequence logo was generated with Weblogo [81] based on a multiple-sequence alignment of 1975 LOV domain sequences for a recently published data set [80]. B Close-up view of PpSB1-LOV with residue backbone positions for residues within (red) and around (violet) the hydrophobic I48 pocket shown as spheres (compare to red and violet asterisks in panel A). C Multiple sequence alignment of the LOV domain of PpSB1-LOV, YtvA, AsLOV2 and three selected putative LOV photoreceptors with a polar substitution at the I48 position of PpSB1-LOV (marked with red asterisk in square brackets). Residues constituting the hydrophobic pocket are marked by red asterisk. Sequence information can be found in the Supporting Information Table S6
Interestingly, while the most common residues that constitute the corresponding pocket in LOV domains are indeed mostly hydrophobic, we still find naturally occurring polar substitution for all positions, albeit with generally low probabilities. The most common polar residue type identified in the data set for the I48 position of PpSB1-LOV is threonine, which is found in 3.2% of the analyzed sequences, followed by serine found in 1.4% of the sequences. Putative LOV photoreceptors containing either a serine or threonine at the position of I48 of PpSB1-LOV include among others a LOV domain containing protein from Coccomyxa subellipsoidea, a LOV histidine kinase of Erythrobacter sp. and a LOV histidine kinase of Pseudomonas syringae (see Fig. 5, C marked by a red asterisk in square brackets). Polar I48 substitutions can, thus, be found in eukaryotes and phototrophic and non-phototrophic prokaryotes, indicative of a more general tuning mechanism that is not restricted to certain genera. A list of selected proteins containing polar I48 substitutions can be found in Supporting Table S6.
Based on these findings, it is tempting to speculate that polar substitutions within the hydrophobic I48 pocket, as the ones we have introduced here artificially by site-directed mutagenesis in PpSB1-LOV, are indeed used in nature to tune the dark-recovery kinetics of LOV domains. Further studies of LOV proteins naturally containing polar residues within the hydrophobic I48 pocket would, however, be needed to corroborate this notion.
5 Conclusions
In conclusion, the here-presented study identifies a conserved, but previously overlooked structural region outside the flavin-binding pocket of LOV domains, for which mutations have a strong impact on the dark-recovery reaction across different LOV photoreceptor families. Moreover, the identified I48T mutation in PpSB1-LOV is structurally and mechanistically benign, i.e., as light-induced structural changes, as probed by NMR spectroscopy and X-ray crystallography, are not altered in the variant. Please note that, due to the lack of a straightforward functional read-out for the here-studied PpSB1-LOV system, the impact of the mutation on the cellular function of the protein could not be tested. In addition, whether or not, the homologous mutation in other LOV photoreceptors, as well as other mutations in the hydrophobic pocket of PpSB1-LOV are also functionally benign, remains to be addressed in future studies. Nevertheless, due to the conserved nature of the corresponding structural region, the here identified mutations should find application in dark-recovery tuning of optogenetic tools and LOV photoreceptors, alike.
Data availability
All datasets generated during and/or analyzed as part of the present study are available from the corresponding author upon reasonable request.
References
Cheng, Z., Yamamoto, H., & Bauer, C. E. (2016). Cobalamin’s (Vitamin B12) surprising function as a photoreceptor. Trends in Biochemical Sciences, 41, 647–650.
Conrad, K. S., Manahan, C. C., & Crane, B. R. (2014). Photochemistry of flavoprotein light sensors. Nature Chemical Biology, 10, 801–809.
Crosson, S., Rajagopal, S., & Moffat, K. (2003). The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry (US), 42, 2–10.
Fushimi, K., & Narikawa, R. (2019). Cyanobacteriochromes: Photoreceptors covering the entire UV-to-visible spectrum. Current Opinion in Structural Biology, 57, 39–46.
Kirilovsky, D., & Kerfeld, C. A. (2013). The orange carotenoid protein: A blue-green light photoactive protein. Photochemical and Photobiological Sciences, 12, 1135–1143.
Möglich, A., & Moffat, K. (2010). Engineered photoreceptors as novel optogenetic tools. Photochemical and Photobiological Sciences, 9, 1286–1300.
Rizzini, L., Favory, J. J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., Baumeister, R., Schafer, E., Nagy, F., Jenkins, G. I., & Ulm, R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science, 332, 103–106.
Schreiber, M., Sugihara, M., Okada, T., & Buss, V. (2006). Quantum mechanical studies on the crystallographic model of bathorhodopsin. Angewandte Chemie International Edition, 45, 4274–4277.
van der Horst, M. A., & Hellingwerf, K. J. (2004). Photoreceptor proteins, “star actors of modern times”: A review of the functional dynamics in the structure of representative members of six different photoreceptor families. Accounts of Chemical Research, 37, 13–20.
Krauss, U., Minh, B. Q., Losi, A., Gärtner, W., Eggert, T., von Haeseler, A., & Jaeger, K. E. (2009). Distribution and phylogeny of light-oxygen-voltage-blue-light-signaling proteins in the three kingdoms of life. Journal of Bacteriology, 191, 7234–7242.
Christie, J. M., Salomon, M., Nozue, K., Wada, M., & Briggs, W. R. (1999). LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proceedings of the National Academy of Sciences of the United States of America, 96, 8779–8783.
Salomon, M., Christie, J. M., Knieb, E., Lempert, U., & Briggs, W. R. (2000). Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry (US), 39, 9401–9410.
Möglich, A., Ayers, R. A., & Moffat, K. (2009). Structure and signaling mechanism of Per-ARNT-Sim domains. Structure, 17, 1282–1294.
Taylor, B. L., & Zhulin, I. B. (1999). PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiology and Molecular Biology Reviews, 63, 479–506.
Suetsugu, N., & Wada, M. (2013). Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: Phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant and Cell Physiology, 54, 8–23.
Huang, K., & Beck, C. F. (2003). Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences of the United States of America, 100, 6269–6274.
Huang, K., Merkle, T., & Beck, C. F. (2002). Isolation and characterization of a Chlamydomonas gene that encodes a putative blue-light photoreceptor of the phototropin family. Physiologia Plantarum, 115, 613–622.
Im, C. S., Eberhard, S., Huang, K., Beck, C. F., & Grossman, A. R. (2006). Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. The Plant Journal, 48, 1–16.
Petroutsos, D., Tokutsu, R., Maruyama, S., Flori, S., Greiner, A., Magneschi, L., Cusant, L., Kottke, T., Mittag, M., Hegemann, P., Finazzi, G., & Minagawa, J. (2016). A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature, 537, 563–566.
Trippens, J., Greiner, A., Schellwat, J., Neukam, M., Rottmann, T., Lu, Y., Kateriya, S., Hegemann, P., & Kreimer, G. (2012). Phototropin influence on eyespot development and regulation of phototactic behavior in Chlamydomonas reinhardtii. The Plant Cell, 24, 4687–4702.
Takahashi, F., Yamagata, D., Ishikawa, M., Fukamatsu, Y., Ogura, Y., Kasahara, M., Kiyosue, T., Kikuyama, M., Wada, M., & Kataoka, H. (2007). AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proceedings of the National academy of Sciences of the United States of America, 104, 19625–19630.
Yu, Z., & Fischer, R. (2019). Light sensing and responses in fungi. Nature Reviews Microbiology, 17, 25–36.
Endres, S., Granzin, J., Circolone, F., Stadler, A., Krauss, U., Drepper, T., Svensson, V., Knieps-Grünhagen, E., Wirtz, A., Cousin, A., Tielen, P., Willbold, D., Jaeger, K. E., & Batra-Safferling, R. (2015). Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae. BMC Microbiology, 15, 30.
Herrou, J., & Crosson, S. (2011). Function, structure and mechanism of bacterial photosensory LOV proteins. Nature Reviews Microbiology, 9, 713–723.
Jentzsch, K., Wirtz, A., Circolone, F., Drepper, T., Losi, A., Gärtner, W., Jaeger, K. E., & Krauss, U. (2009). Mutual exchange of kinetic properties by extended mutagenesis in two short LOV domain proteins from Pseudomonas putida. Biochemistry (US), 48, 10321–10333.
Krauss, U., Losi, A., Gärtner, W., Jaeger, K. E., & Eggert, T. (2005). Initial characterization of a blue-light sensing, phototropin-related protein from Pseudomonas putida: A paradigm for an extended LOV construct. Physical Chemistry Chemical Physics, 7, 2804–2811.
Röllen, K., Granzin, J., Batra-Safferling, R., & Stadler, A. M. (2018). Small-angle X-ray scattering study of the kinetics of light-dark transition in a LOV protein. PLoS ONE, 13, e0200746.
Röllen, K., Granzin, J., Panwalkar, V., Arinkin, V., Rani, R., Hartmann, R., Krauss, U., Jaeger, K. E., Willbold, D., & Batra-Safferling, R. (2016). Signaling states of a short blue-light photoreceptor protein PpSB1-LOV revealed from crystal structures and solution NMR spectroscopy. Journal of Molecular Biology, 428, 3721–3736.
Stadler, A. M., Knieps-Grünhagen, E., Bocola, M., Lohstroh, W., Zamponi, M., & Krauss, U. (2016). Photoactivation reduces side-chain dynamics of a LOV photoreceptor. Biophysical Journal, 110, 1064–1074.
Conrad, K. S., Bilwes, A. M., & Crane, B. R. (2013). Light-induced subunit dissociation by a light-oxygen-voltage domain photoreceptor from Rhodobacter sphaeroides. Biochemistry (US), 52, 378–391.
Schwerdtfeger, C., & Linden, H. (2003). VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO Journal, 22, 4846–4855.
Chen, C. H., DeMay, B. S., Gladfelter, A. S., Dunlap, J. C., & Loros, J. J. (2010). Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proceedings of the National academy of Sciences of the United States of America, 107, 16715–16720.
Sumi, S., Mutaguchi, N., Ebuchi, T., Tsuchida, H., Yamamoto, T., Suzuki, M., Natsuka, C., Shiratori-Takano, H., Shintani, M., Nojiri, H., Ueda, K., & Takano, H. (2020). Light Response of Pseudomonas putida KT2440 Mediated by Class II LitR, a Photosensor Homolog. Journal of Bacteriology, 202, e00146-e220.
Vaidya, A. T., Chen, C. H., Dunlap, J. C., Loros, J. J., & Crane, B. R. (2011). Structure of a light-activated LOV protein dimer that regulates transcription. Science Signaling, 4, ra50.
Losi, A., Gardner, K. H., & Möglich, A. (2018). Blue-light receptors for optogenetics. Chemical Reviews, 118, 10659–10709.
Hart, J. E., Sullivan, S., Hermanowicz, P., Petersen, J., Diaz-Ramos, L. A., Hoey, D. J., Labuz, J., & Christie, J. M. (2019). Engineering the phototropin photocycle improves photoreceptor performance and plant biomass production. Proceedings of the National academy of Sciences of the United States of America, 116, 12550–12557.
Ziegler, T., Schumacher, C. H., & Möglich, A. (2016). Guidelines for photoreceptor engineering. Methods in Molecular Biology, 1408, 389–403.
Kogler, A. C., Kherdjemil, Y., Bender, K., Rabinowitz, A., Marco-Ferreres, R., & Furlong, E. E. M. (2021). Extremely rapid and reversible optogenetic perturbation of nuclear proteins in living embryos. Developmental Cell, 56(2348–2363), e8.
Forlani, G., Antwi, E. B., Weis, D., Ozturk, M. A., Queck, B. A. W., Brecht, D., & Di Ventura, B. (2022). Analysis of slow-cycling variants of the light-inducible nuclear protein export system LEXY in mammalian Cells. ACS Synthetic Biology, 11, 3529–3533.
Spiltoir, J. I., Strickland, D., Glotzer, M., & Tucker, C. L. (2016). Optical control of peroxisomal trafficking. ACS Synthetic Biology, 5, 554–560.
Kottke, T., Heberle, J., Hehn, D., Dick, B., & Hegemann, P. (2003). Phot-LOV1: Photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophysical Journal, 84, 1192–1201.
Song, S. H., Freddolino, P. L., Nash, A. I., Carroll, E. C., Schulten, K., Gardner, K. H., & Larsen, D. S. (2011). Modulating LOV domain photodynamics with a residue alteration outside the chromophore binding site. Biochemistry (US), 50, 2411–2423.
Song, S. H., Madsen, D., van der Steen, J. B., Pullman, R., Freer, L. H., Hellingwerf, K. J., & Larsen, D. S. (2013). Primary photochemistry of the dark and light-adapted states of the YtvA protein from bacillus subtilis. Biochemistry (US), 52, 7951–7963.
Swartz, T. E., Corchnoy, S. B., Christie, J. M., Lewis, J. W., Szundi, I., Briggs, W. R., & Bogomolni, R. A. (2001). The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. Journal of Biological Chemistry, 276, 36493–36500.
Zhu, J., Mathes, T., Hontani, Y., Alexandre, M. T., Toh, K. C., Hegemann, P., & Kennis, J. T. (2016). Photoadduct formation from the FMN singlet excited state in the LOV2 domain of Chlamydomonas reinhardtii phototropin. Journal of Physical Chemistry Letters, 7, 4380–4384.
Arinkin, V., Granzin, J., Röllen, K., Krauss, U., Jaeger, K. E., Willbold, D., & Batra-Safferling, R. (2017). Structure of a LOV protein in apo-state and implications for construction of LOV-based optical tools. Scientific Reports, 7, 42971.
Circolone, F., Granzin, J., Jentzsch, K., Drepper, T., Jaeger, K. E., Willbold, D., Krauss, U., & Batra-Safferling, R. (2012). Structural basis for the slow dark recovery of a full-length LOV protein from Pseudomonas putida. Journal of Molecular Biology, 417, 362–374.
Pudasaini, A., Shim, J. S., Song, Y. H., Shi, H., Kiba, T., Somers, D. E., Imaizumi, T., & Zoltowski, B. D. (2017). Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis. eLife. https://doi.org/10.7554/eLife.21646
Losi, A., Polverini, E., Quest, B., & Gärtner, W. (2002). First evidence for phototropin-related blue-light receptors in prokaryotes. Biophysical Journal, 82, 2627–2634.
Nakasone, Y., Zikihara, K., Tokutomi, S., & Terazima, M. (2010). Kinetics of conformational changes of the FKF1-LOV domain upon photoexcitation. Biophysical Journal, 99, 3831–3839.
Diensthuber, R. P., Engelhard, C., Lemke, N., Gleichmann, T., Ohlendorf, R., Bittl, R., & Möglich, A. (2014). Biophysical, mutational, and functional investigation of the chromophore-binding pocket of light-oxygen-voltage photoreceptors. ACS Synthetic Biology, 3, 811–819.
Fettweiss, T., Röllen, K., Granzin, J., Reiners, O., Endres, S., Drepper, T., Willbold, D., Jaeger, K. E., Batra-Safferling, R., & Krauss, U. (2018). Mechanistic Basis of the fast dark recovery of the short LOV Protein DsLOV from Dinoroseobacter shibae. Biochemistry (US), 57, 4833–4847.
Zayner, J. P., Antoniou, C., French, A. R., Hause, R. J., Jr., & Sosnick, T. R. (2013). Investigating models of protein function and allostery with a widespread mutational analysis of a light-activated protein. Biophysical Journal, 105, 1027–1036.
Dietler, J., Gelfert, R., Kaiser, J., Borin, V., Renzl, C., Pilsl, S., Ranzani, A. T., Garcia de Fuentes, A., Gleichmann, T., Diensthuber, R. P., Weyand, M., Mayer, G., Schapiro, I., & Möglich, A. (2022). Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine. Nature Communications, 13, 2618.
Arinkin, V., Granzin, J., Krauss, U., Jaeger, K. E., Willbold, D., & Batra-Safferling, R. (2021). Structural determinants underlying the adduct lifetime in the LOV proteins of Pseudomonas putida. FEBS Journal, 288, 4955–4972.
Raffelberg, S., Mansurova, M., Gärtner, W., & Losi, A. (2011). Modulation of the photocycle of a LOV domain photoreceptor by the hydrogen-bonding network. Journal of the American Chemical Society, 133, 5346–5356.
Zoltowski, B. D., Nash, A. I., & Gardner, K. H. (2011). Variations in protein-flavin hydrogen bonding in a light, oxygen, voltage domain produce non-Arrhenius kinetics of adduct decay. Biochemistry (US), 50, 8771–8779.
Christie, J. M., Corchnoy, S. B., Swartz, T. E., Hokenson, M., Han, I. S., Briggs, W. R., & Bogomolni, R. A. (2007). Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry (US), 46, 9310–9319.
Zoltowski, B. D., Vaccaro, B., & Crane, B. R. (2009). Mechanism-based tuning of a LOV domain photoreceptor. Nature Chemical Biology, 5, 827–834.
Brosi, R., Illarionov, B., Mathes, T., Fischer, M., Joshi, M., Bacher, A., Hegemann, P., Bittl, R., Weber, S., & Schleicher, E. (2010). Hindered rotation of a cofactor methyl group as a probe for protein-cofactor interaction. Journal of the American Chemical Society, 132, 8935–8944.
Alexandre, M. T., Arents, J. C., van Grondelle, R., Hellingwerf, K. J., & Kennis, J. T. (2007). A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry (US), 46, 3129–3137.
Pennacchietti, F., Abbruzzetti, S., Losi, A., Mandalari, C., Bedotti, R., Viappiani, C., Zanacchi, F. C., Diaspro, A., & Gartner, W. (2014). The dark recovery rate in the photocycle of the bacterial photoreceptor YtvA is affected by the cellular environment and by hydration. PLoS ONE, 9, e107489.
Purcell, E. B., McDonald, C. A., Palfey, B. A., & Crosson, S. (2010). An analysis of the solution structure and signaling mechanism of LovK, a sensor histidine kinase integrating light and redox signals. Biochemistry (US), 49, 6761–6770.
Kawano, F., Aono, Y., Suzuki, H., & Sato, M. (2013). Fluorescence imaging-based high-throughput screening of fast- and slow-cycling LOV proteins. PLoS ONE, 8, e82693.
Buttani, V., Losi, A., Eggert, T., Krauss, U., Jaeger, K. E., Cao, Z., & Gärtner, W. (2007). Conformational analysis of the blue-light sensing protein YtvA reveals a competitive interface for LOV-LOV dimerization and interdomain interactions. Photochemical and Photobiological Sciences, 6, 41–49.
Kaschner, M., Loeschcke, A., Krause, J., Minh, B. Q., Heck, A., Endres, S., Svensson, V., Wirtz, A., von Haeseler, A., Jaeger, K. E., Drepper, T., & Krauss, U. (2014). Discovery of the first light-dependent protochlorophyllide oxidoreductase in anoxygenic phototrophic bacteria. Molecular Microbiology, 93, 1066–1078.
Findeisen, M., Brand, T., & Berger, S. (2007). A 1H-NMR thermometer suitable for cryoprobes. Magnetic Resonance in Chemistry, 45, 175–178.
Grzesiek, S., & Bax, A. (1992). Improved 3d triple-resonance Nmr techniques applied to a 31-Kda protein. Journal of Magnetic Resonance, 96, 432–440.
Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., & Bax, A. (1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR, 6, 277–293.
Vranken, W. F., Boucher, W., Stevens, T. J., Fogh, R. H., Pajon, A., Llinas, M., Ulrich, E. L., Markley, J. L., Ionides, J., & Laue, E. D. (2005). The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins, 59, 687–696.
Grzesiek, S., Stahl, S. J., Wingfield, P. T., & Bax, A. (1996). The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef Mapping of the Nef binding surface by NMR. Biochemistry (US), 35, 10256–10261.
de Sanctis, D., Beteva, A., Caserotto, H., Dobias, F., Gabadinho, J., Giraud, T., Gobbo, A., Guijarro, M., Lentini, M., Lavault, B., Mairs, T., McSweeney, S., Petitdemange, S., Rey-Bakaikoa, V., Surr, J., Theveneau, P., Leonard, G. A., & Mueller-Dieckmann, C. (2012). ID29: A high-intensity highly automated ESRF beamline for macromolecular crystallography experiments exploiting anomalous scattering. Journal of Synchrotron Radiation, 19, 455–461.
Nurizzo, D., Mairs, T., Guijarro, M., Rey, V., Meyer, J., Fajardo, P., Chavanne, J., Biasci, J. C., McSweeney, S., & Mitchell, E. (2006). The ID23-1 structural biology beamline at the ESRF. Journal of Synchrotron Radiation, 13, 227–238.
Bourenkov, G. P., & Popov, A. N. (2010). Optimization of data collection taking radiation damage into account. Acta Crystallographica D, 66, 409–419.
Kabsch, W. (2010). Xds. Acta Crystallographica Section D-Biological Crystallography, 66, 125–132.
Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A., & Wilson, K. S. (2011). Overview of the CCP4 suite and current developments. Acta Crystallographica Section D Biological Crystallography, 67, 235–242.
Kantardjieff, K. A., & Rupp, B. (2003). Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins DNA, and protein-nucleic acid complex crystals. Protein Science, 12, 1865–1871.
Emsley, P., & Cowtan, K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallographica Section D Biological Crystallography, 60, 2126–2132.
Kopka, B., Magerl, K., Savitsky, A., Davari, M. D., Röllen, K., Bocola, M., Dick, B., Schwaneberg, U., Jaeger, K. E., & Krauss, U. (2017). Electron transfer pathways in a light, oxygen, voltage (LOV) protein devoid of the photoactive cysteine. Scientific Reports, 7, 1–6.
Glantz, S. T., Carpenter, E. J., Melkonian, M., Gardner, K. H., Boyden, E. S., Wong, G. K., & Chow, B. Y. (2016). Functional and topological diversity of LOV domain photoreceptors. Proceedings of the National Academy of Sciences of the United States of America, 113, E1442–E1451.
Crooks, G. E., Hon, G., Chandonia, J. M., & Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Research, 14, 1188–1190.
Wheeler, T. J., Clements, J., & Finn, R. D. (2014). Skylign: A tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics, 15, 7.
Pudasaini, A., El-Arab, K. K., & Zoltowski, B. D. (2015). LOV-based optogenetic devices: Light-driven modules to impart photoregulated control of cellular signaling. Frontiers in Molecular Biosciences, 2, 18.
Ziegler, T., & Möglich, A. (2015). Photoreceptor engineering. Frontiers in Molecular Biosciences, 2, 30.
Acknowledgements
This work was supported by grants from the Federal Ministry of Education and Research (Project OptoSys, FKZ 031A16) and by the “Vernetzungsdoktorandenförderung” of the Helmholtz Association. The X-ray diffraction experiments were performed on the beamlines ID23-1 and ID29 at the ESRF (Grenoble, France). We are grateful to the local contact scientists at ESRF for providing help in using the beamline.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
UK, VP and RB-S conceived the study. SH generated constructs, purified the proteins for spectroscopic and X-ray crystallographic studies, performed UV/Vis spectroscopic analyses and analyzed the corresponding data. MS and VP performed NMR measurements, analyzed and interpreted NMR data. E K-G helped with protein purification and prepared proteins samples for NMR measurements. JG collected X-ray diffraction data and solved the structure of the I48T variant. JG and RB-S analyzed and interpreted the obtained structural data. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests that are relevant to the content of this article.
Additional information
This publication is dedicated to Prof. Silvia E. Braslavsky, a pioneer in photobiology and photobiophysics, on the occasion of her 80th birthday.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hemmer, S., Schulte, M., Knieps-Grünhagen, E. et al. Residue alterations within a conserved hydrophobic pocket influence light, oxygen, voltage photoreceptor dark recovery. Photochem Photobiol Sci 22, 713–727 (2023). https://doi.org/10.1007/s43630-022-00346-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43630-022-00346-5