Abstract
Background
Lumbar disc herniation is a common degenerative lumbar disease with an increasing incidence. Percutaneous endoscopic lumbar discectomy can treat lumbar disc herniation safely and effectively with a minimally invasive procedure. However, the learning curve of this technology is steep, which means that initial learners are often not sufficiently proficient in endoscopic operations, which can easily lead to iatrogenic damage. At present, the application of computer deep learning technology to clinical diagnosis, treatment, and surgical navigation has achieved satisfactory results.
Purpose
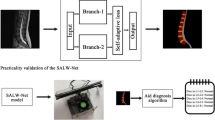
The objective of our team is to develop a multi-element identification system for the visual field of endoscopic spine surgery using deep learning algorithms and to evaluate the feasibility of this system.
Method
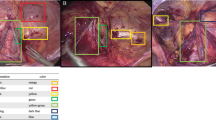
We established an image database by collecting surgical videos of 48 patients diagnosed with lumbar disc herniation, which was labeled by two spinal surgeons. We selected 6000 images of the visual field of percutaneous endoscopic spine surgery (including various tissue structures and surgical instruments), divided into the training data, validation data, and test data according to 2:1:2. We developed convolutional neural network models based on instance segmentation—Solov2, CondInst, Mask R-CNN and Yolact, and set the four network model backbone as ResNet101 and ResNet50 respectively. Mean average precision (mAP) and frames per second (FPS) were used to measure the performance of each model for classification, localization and recognition in real time, and AP (average) is used to evaluate how easily an element is detected by neural networks based on computer deep learning.
Result
Comprehensively comparing mAP and FSP of each model for bounding box test and segmentation task for the test set of images, we found that Solov2 (ResNet101) (mAP = 73.5%, FPS = 28.9), Mask R-CNN (ResNet101) (mAP = 72.8%, FPS = 28.5) models are the most stable, with higher precision and faster image processing speed. Combining the average precision of the elements in the bounding box test and segmentation tasks in each network, the AP(average) was highest for tool 3 (bbox-0.85, segm-0.89) and lowest for tool 5 (bbox-0.63, segm-0.72) in the instrumentation, whereas in the anatomical tissue elements, the fibrosus annulus (bbox-0.68, segm-0.69) and ligamentum flavum (bbox-0.65, segm-0.62) had higher AP(average),while extra-dural fat (bbox-0.42, segm-0.44) was lowest.
Conclusion
Our team has developed a multi-element identification system for the visual field of percutaneous endoscopic spine surgery adapted to the interlaminar and foraminal approaches, which can identify and track anatomical tissue (nerve, ligamentum flavum, nucleus pulposus, etc.) and surgical instruments (endoscopic forceps, an high-speed diamond burr, etc.), which can be used in the future as a virtual educational tool or applied to the intraoperative real-time assistance system for spinal endoscopic operation.



Similar content being viewed by others
Availability of Data and Materials
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
References:
Zhang, A. S., Xu, A., Ansari, K., Hardacker, K., Anderson, G., Alsoof, D., & Daniels, A. H. (2023). Lumbar Disc Herniation: Diagnosis and Management. American Journal of Medicine, 136, 645–651. https://doi.org/10.1016/j.amjmed.2023.03.024
Shen, S. C., Chen, H. C., Tsou, H. K., Lin, R. H., Shih, Y. T., Huang, C. W., Tang, C. L., Chen, H. T., Chang, C. C., & Tzeng, C. Y. (2023). Percutaneous endoscopic lumbar discectomy for L5–S1 disc herniation based on image analysis and clinical findings: A retrospective review of 345 cases. Medicine (Baltimore), 102, e32832. https://doi.org/10.1097/MD.0000000000032832
Pan, M., Li, Q., Li, S., Mao, H., Meng, B., Zhou, F., & Yang, H. (2020). Percutaneous endoscopic lumbar discectomy: Indications and complications. Pain Physician, 23, 49–56.
Cheng, Y. P., Cheng, X. K., & Wu, H. (2022). A comparative study of percutaneous endoscopic interlaminar discectomy and transforaminal discectomy for L5–S1 calcified lumbar disc herniation. BMC Musculoskeletal Disorders, 23, 244. https://doi.org/10.1186/s12891-022-05186-z
Ahn, Y., Lee, S., Son, S., Kim, H., & Kim, J. E. (2020). Learning curve for transforaminal percutaneous endoscopic lumbar discectomy: A systematic review. World Neurosurgery, 143, 471–479. https://doi.org/10.1016/j.wneu.2020.08.044
Ao, S., Wu, J., Tang, Y., Zhang, C., Li, J., Zheng, W., & Zhou, Y. (2019). Percutaneous endoscopic lumbar discectomy assisted by O-arm-based navigation improves the learning curve. BioMed Research International, 2019, 6509409. https://doi.org/10.1155/2019/6509409
Hosny, A., Parmar, C., Quackenbush, J., Schwartz, L. H., & Aerts, H. (2018). Artificial intelligence in radiology. Nature Reviews Cancer, 18, 500–510. https://doi.org/10.1038/s41568-018-0016-5
Yeh, Y. C., Weng, C. H., Huang, Y. J., Fu, C. J., Tsai, T. T., & Yeh, C. Y. (2021). Deep learning approach for automatic landmark detection and alignment analysis in whole-spine lateral radiographs. Science and Reports, 11, 7618. https://doi.org/10.1038/s41598-021-87141-x
Fan, N., Yuan, S., Du, P., Zhu, W., Li, L., Hai, Y., Ding, H., Wang, G., & Zang, L. (2020). Design of a robot-assisted system for transforaminal percutaneous endoscopic lumbar surgeries: Study protocol. Journal of Orthopaedic Surgery and Research, 15, 479. https://doi.org/10.1186/s13018-020-02003-y
Hagan, M. J., Remacle, T., Leary, O. P., Feler, J., Shaaya, E., Ali, R., Zheng, B., Bajaj, A., Traupe, E., Kraus, M., et al. (2022). Navigation Techniques in Endoscopic Spine Surgery. BioMed Research International, 2022, 8419739. https://doi.org/10.1155/2022/8419739
Fan, N., Yuan, S., Du, P., Wu, Q., Wang, T., Wang, A., Li, J., Kong, X., Zhu, W., & Zang, L. (2021). Complications and risk factors of percutaneous endoscopic transforaminal discectomy in the treatment of lumbar spinal stenosis. BMC Musculoskeletal Disorders, 22, 1041. https://doi.org/10.1186/s12891-021-04940-z
Fan, G., Liu, H., Wang, D., Feng, C., Li, Y., Yin, B., Zhou, Z., Gu, X., Zhang, H., Lu, Y., et al. (2020). Deep learning-based lumbosacral reconstruction for difficulty prediction of percutaneous endoscopic transforaminal discectomy at L5/S1 level: A retrospective cohort study. International Journal of Surgery, 82, 162–169. https://doi.org/10.1016/j.ijsu.2020.08.036
Cui, P., Shu, T., Lei, J., & Chen, W. (2021). Nerve recognition in percutaneous transforaminal endoscopic discectomy using convolutional neural network. Medical Physics, 48, 2279–2288. https://doi.org/10.1002/mp.14822
Cho, S. M., Kim, Y. G., Jeong, J., Kim, I., Lee, H. J., & Kim, N. (2021). Automatic tip detection of surgical instruments in biportal endoscopic spine surgery. Computers in Biology and Medicine, 133, 104384. https://doi.org/10.1016/j.compbiomed.2021.104384
Lokhande, P. V. (2023). Full endoscopic spine surgery. Journal of Orthopaedics, 40, 74–82. https://doi.org/10.1016/j.jor.2023.04.010
Zhang, Y., Chu, J., Leng, L., & Miao, J. (2020). Mask-refined R-CNN: a network for refining object details in instance segmentation. Sensors (Basel). https://doi.org/10.3390/s20041010
He, K., Gkioxari, G., Dollar, P., & Girshick, R. (2020). Mask R-CNN. IEEE Transactions on Pattern Analysis and Machine Intelligence, 42, 386–397. https://doi.org/10.1109/TPAMI.2018.2844175
Wang, N., Zhang, J., & Song, X. (2023). A pipeline defect instance segmentation system based on SparseInst. Sensors (Basel). https://doi.org/10.3390/s23229019
Bolya, D., Zhou, C., Xiao, F., & Lee, Y. J. (2022). YOLACT++ better real-time instance segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 44, 1108–1121. https://doi.org/10.1109/TPAMI.2020.3014297
Yu, Z., Liu, L., Jiao, H., Chen, J., Chen, Z., Song, Z., Lin, H., & Tian, F. (2022). Leveraging SOLOv2 model to detect heat stress of poultry in complex environments. Frontiers in Veterinary Science, 9, 1062559. https://doi.org/10.3389/fvets.2022.1062559
Alaeddine, H., & Jihene, M. (2021). Deep residual network in network. Computational Intelligence and Neuroscience, 2021, 6659083. https://doi.org/10.1155/2021/6659083
Reiner, A. J., Hollands, J. G., & Jamieson, G. A. (2017). Target detection and identification performance using an automatic target detection system. Human Factors, 59, 242–258. https://doi.org/10.1177/0018720816670768
Zarvani, M., Saberi, S., Azmi, R., & Shojaedini, S. V. (2021). Residual learning: a new paradigm to improve deep learning-based segmentation of the left ventricle in magnetic resonance imaging cardiac images. Journal of Medical Signals and Sensors, 11, 159–168. https://doi.org/10.4103/jmss.JMSS_38_20
Acknowledgements
Thank you for all the support from the XuZhou Central Hospital and Xuzhou Medical University.
Funding
This study was supported by the Projects of Medical Key and leading talents of Xuzhou (XWRCHT20220050, XWRCHT20210035), Xuzhou Plan of introducing a team of clinical medical experts(2019TD002), the Project of Health Innovation Teams of Xuzhou (XWCX201601), Xuzhou Medical Foundation for Youth Reserved Experts Fund (2014006), Jiangsu Provincial Medical Youth Talent (QNRC2016392), the Scientific Research Projects of Jiangsu Provincial Health Commission (M2022048, Z2022040), “Six one Projects” for High-level Health Talents in Jiangsu Province (LGY2018047), Jiangsu Province “333” talents Project, The Project of Science and Technology of Xuzhou (KC22161), Postdoctoral Project of China (314233).
Author information
Authors and Affiliations
Contributions
JHB, YRW, JQZ and GWL contributed to conception and design of the study. JHB, SH, JL, ZFW and LX organized the database. JHB, YL, BH, MHD and GPL performed the statistical analysis. JHB, RN, CM and GWL wrote the first draft of the manuscript. JHB, YL, CM were involved in article revisions. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Review Committee of Xuzhou Central Hospital under the ethical number XZXY-LK-20221020-095, in accordance with the Declaration of Helsinki. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Consent for Publication
All authors contributed to the article and approved the submitted version.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bu, J., Lei, Y., Wang, Y. et al. A Multi-Element Identification System Based on Deep Learning for the Visual Field of Percutaneous Endoscopic Spine Surgery. JOIO 58, 587–597 (2024). https://doi.org/10.1007/s43465-024-01134-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43465-024-01134-2