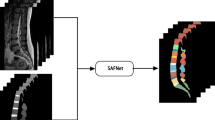
Abstract
Segmentation of intervertebral discs and vertebrae from spine magnetic resonance (MR) images is essential to aid diagnosis algorithms for lumbar disc herniation. Convolutional neural networks (CNN) are effective methods, but often require high computational costs. Designing a lightweight CNN is more suitable for medical sites lacking high-computing power devices, yet due to the unbalanced pixel distribution in spine MR images, the segmentation is often sub-optimal. To address this issue, a lightweight spine segmentation CNN based on a self-adjusting loss function, which is named SALW-Net, is proposed in this study. For SALW-Net, the self-adjusting loss function could dynamically adjust the loss weights of the two branches according to the differences in segmentation results and labels during the training; thus, the ability for learning unbalanced pixels is enhanced. Two separate datasets are used to evaluate the proposed SALW-Net. Specifically, the proposed SALW-Net has fewer parameter numbers than U-net (only 2%) but achieves higher evaluation scores than that of U-net (the average DSC score of SALW-Net is 0.8781, and that of U-net is 0.8482). In addition, the practicality validation for SALW-Net is also proceeding, including deploying the model on a lightweight device and producing an aid diagnosis algorithm based on segmentation results. This means our SALW-Net has clinical application potential for assisted diagnosis in low computational power scenarios.
Graphical Abstract













Similar content being viewed by others
References
Feng Y, Egan B, Wang J (2016) Genetic factors in intervertebral disc degeneration. Genes Dis 3:178–185. https://doi.org/10.1016/j.gendis.2016.04.005
Zhang Q, Chon T, Zhang Y, Baker JS, Gu Y (2021) Finite element analysis of the lumbar spine in adolescent idiopathic scoliosis subjected to different loads. Comput Biol Med 136:104745. https://doi.org/10.1016/j.compbiomed.2021.104745
Özcan F, Alkan A (2021) Frontal cortex neuron type classification with deep learning and recurrence plot. Traitement Du Signal 38:807–819. https://doi.org/10.18280/ts.380327
Bakator M, Radosav D (2018) Deep learning and medical diagnosis: a review of literature. Multimodal Technol Interact 2(3):47. https://doi.org/10.3390/mti2030047
Hwang E-J, Kim S, Jung J-Y (2022) Fully automated segmentation of lumbar bone marrow in sagittal, high-resolution T1-weighted magnetic resonance images using 2D U-NET. Comput Biol Med 140:105105. https://doi.org/10.1016/j.compbiomed.2021.105105
Dolz J, Desrosiers C, Ben Ayed I. IVD-Net: intervertebral disc localization and segmentation in MRI with a multi-modal UNet. Proc. Computational Methods and Clinical Applications for Spine Imaging: 5th International Workshop and Challenge, CSI 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16, 2018, Revised Selected Papers, 2019:130-143. https://doi.org/10.1007/978-3-030-13736-6_11
Wang C, Guo Y, Chen W, Yu Z (2019) Fully automatic intervertebral disc segmentation using multimodal 3D U-Net. Proc. 2019 IEEE 43rd Annual Computer Software and Applications Conference (COMPSAC). Proc 2019 IEEE 43rd Ann Comput Software Appl Conference (COMPSAC) 1:730–739. https://doi.org/10.1109/COMPSAC.2019.00109
Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y et al (2021) Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data 8:1–74. https://doi.org/10.1186/s40537-021-00444-8
Liu S, Kong W, Chen X, Xu M, Yasir M et al (2022) Multi-scale ship detection algorithm based on a lightweight neural network for spaceborne SAR images. Remote Sens-Basel 14(5):1149. https://doi.org/10.3390/rs14051149
Zhang R, Zhu F, Liu J, Liu G (2019) Depth-wise separable convolutions and multi-level pooling for an efficient spatial CNN-based steganalysis. IEEE T Inf Foren Sec 15:1138–1150. https://doi.org/10.1109/TIFS.2019.2936913
Park J, Woo S, Lee J-Y, Kweon IS (2018) Bam: bottleneck attention module. arXiv preprint arXiv:1807.06514. https://doi.org/10.48550/arXiv.1807.06514
Zhao H, Qi X, Shen X, Shi J, Jia J (2018) Icnet for real-time semantic segmentation on high-resolution images, Proceedings of the European conference on computer vision (ECCV)2018), 2018:405–420. https://doi.org/10.1007/978-3-030-01219-9_25
Li H, Xiong P, Fan H, Sun J (2019) Dfanet: deep feature aggregation for real-time semantic segmentation, Proceedings of the IEEE/CVF conference on computer vision and pattern recognition2019), 2019:9522–9531. https://doi.org/10.1109/CVPR.2019.00975
Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, et al (2017) Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861. https://doi.org/10.48550/arXiv.1704.04861
Zhao H, Shi J, Qi X, Wang X, Jia J (2017) Pyramid scene parsing network, Proceedings of the IEEE conference on computer vision and pattern recognition2017), 2017:2881-2890. https://doi.org/10.1109/CVPR.2017.660
Paszke A, Chaurasia A, Kim S, Culurciello E (2016) Enet: a deep neural network architecture for real-time semantic segmentation. arXiv preprint arXiv:1606.02147. https://doi.org/10.48550/arXiv.1606.02147
Chen W, Zhang Y, He J, Qiao Y, Chen Y, et al (2019) Prostate segmentation using 2D bridged U-net. Proc. 2019 International Joint Conference on Neural Networks (IJCNN), 2019:1-7. https://doi.org/10.1109/IJCNN.2019.8851908
Jha D, Riegler MA, Johansen D, Halvorsen P, Johansen HD (2020)Doubleu-net: a deep convolutional neural network for medical image segmentation, 2020 IEEE 33rd International symposium on computer-based medical systems (CBMS), pp 558–564. https://doi.org/10.1109/CBMS49503.2020.00111
Cordts M, Omran M, Ramos S, Rehfeld T, Enzweiler M, et al (2016) The cityscapes dataset for semantic urban scene understanding, Proceedings of the IEEE conference on computer vision and pattern recognition2016), pp 3213–3223. https://doi.org/10.1109/CVPR.2016.350
Lin T-Y, Goyal P, Girshick R, He K, Dollár P (2017) Focal loss for dense object detection. Proc. Proceedings of the IEEE international conference on computer vision, 2017:2980-2988https://doi.org/10.1109/TPAMI.2018.2858826
Guo S, Wang K, Kang H, Liu T, Gao Y, Li T (2020) Bin loss for hard exudates segmentation in fundus images. Neurocomputing 392:314–324. https://doi.org/10.1016/j.neucom.2018.10.103
De Brabandere B, Neven D, Van Gool L (2017) Semantic instance segmentation with a discriminative loss function. arXiv preprint arXiv:1708.02551. https://doi.org/10.48550/arXiv.1708.02551
Wang Q, Ma Y, Zhao K, Tian Y (2020) A comprehensive survey of loss functions in machine learning. Ann Data Sci:1–26. https://doi.org/10.1007/s40745-020-00253-5
Garcia-Garcia A, Orts-Escolano S, Oprea S, Villena-Martinez V, Martinez-Gonzalez P, Garcia-Rodriguez J (2018) A survey on deep learning techniques for image and video semantic segmentation. Appl Soft Comput 70:41–65. https://doi.org/10.1016/j.asoc.2018.05.018
Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge Cardoso M (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. Proc. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 14, Proceedings 3, 2017:240–248. https://doi.org/10.1007/978-3-319-67558-9_28
Jafari M, Li R, Xing Y, Auer D, Francis S et al (2019) FU-net: multi-class image segmentation using feedback weighted U-net. Proc. Image and Graphics: 10th International Conference, ICIG 2019, Beijing, China, August 23–25, 2019, Proceedings, Part II 10, 2019:529–537. https://doi.org/10.1007/978-3-030-34110-7_44
Milletari F, Navab N, Ahmadi S-A. (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. Proc. 2016 fourth international conference on 3D vision (3DV), 2016:565–571. https://doi.org/10.1109/3DV.2016.79
Taghanaki SA, Zheng Y, Zhou SK, Georgescu B, Sharma P et al (2019) Combo loss: handling input and output imbalance in multi-organ segmentation. Comput Med Imag Grap 75:24–33. https://doi.org/10.1016/j.compmedimag.2019.04.005
Ma J, Chen J, Ng M, Huang R, Li Y et al (2021) Loss odyssey in medical image segmentation. Med Image Anal 71:102035. https://doi.org/10.1016/j.media.2021.102035
Pang S, Pang C, Zhao L, Chen Y, Su Z et al (2020) SpineParseNet: spine parsing for volumetric MR image by a two-stage segmentation framework with semantic image representation. IEEE T Med Imaging 40:262–273. https://doi.org/10.1109/TMI.2020.3025087
Tuncer SA, Alkan A (2022) Classification of EMG signals taken from arm with hybrid CNN-SVM architecture. Concurr Comp-Pract E 34(5):e6746. https://doi.org/10.1002/cpe.6746
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. Proc. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18, 2015:234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2019) Unet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE T Med Imaging 39:1856–1867. https://doi.org/10.1109/TMI.2019.2959609
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE T Pattern Anal 39:2481–2495. https://doi.org/10.1109/TPAMI.2016.2644615
Sunnetci KM, Kaba E, Celiker FB, Alkan A (2023) Deep network-based comprehensive parotid gland tumor detection. Acad Radiol. https://doi.org/10.1016/j.acra.2023.04.028
Zhang X, Zhou X, Lin M, Sun J (2018) Shufflenet: an extremely efficient convolutional neural network for mobile devices. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp 6848–6856. https://doi.org/10.1109/CVPR.2018.00716
Chen J, Jiang J, Guo X, Tan L (2021) A self-adaptive CNN with PSO for bearing fault diagnosis. Syst Sci Control Eng 9(1):11–22. https://doi.org/10.1080/21642583.2020.1860153
Nandhini S, Ashokkumar K (2021) Improved crossover based monarch butterfly optimization for tomato leaf disease classification using convolutional neural network. Multimed Tools Appl 80:18583–18610. https://doi.org/10.1007/s11042-021-10599-4
Wu H, Zhang B, Liu N (2022) Self-adaptive denoising net: self-supervised learning for seismic migration artifacts and random noise attenuation. J Petrol Sci Eng 214:110431. https://doi.org/10.1016/j.petrol.2022.110431
Serai SD (2022) Basics of magnetic resonance imaging and quantitative parameters T1, T2, T2*, T1rho and diffusion-weighted imaging. Pediatr Radiol 52:217–227. https://doi.org/10.1007/s00247-021-05042-7
Acknowledgements
This work was supported by the Jilin Scientific and Technological Development Program under grant number 20230203098SF.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, S., Li, Q., Li, X. et al. SALW-Net: a lightweight convolutional neural network based on self-adjusting loss function for spine MR image segmentation. Med Biol Eng Comput 62, 1247–1264 (2024). https://doi.org/10.1007/s11517-023-02963-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02963-3