Abstract
Purpose of Review
Gastrointestinal cancers are a major health concern worldwide and include oesophageal, gastric, colorectal, liver, and pancreatic cancers that may have unfavourable patient prognoses and frequently lead to death due to cancer. The function of cancer stem cells (CSCs) in tumour growth, propagation, migration, metastasis, and recurrence has been reported in the literature, and finding effective markers for these cells is highly desirable.
Recent Findings
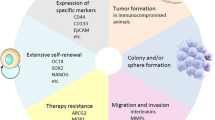
These cells may display properties such as self-renewal, symmetric and asymmetric divisions, low reactive oxygen species, efflux of compounds and toxins, heightened capacity for DNA damage repair, and tumour growth and propagation in immunodeficient mouse models. Furthermore, CSCs dynamically interact with the tumour microenvironment and, in addition, may impact processes like tumour progression, aggressive behaviour, migration, disease relapse, and recurrence, hence becoming clinically important.
Summary
Based on this background, this review aimed to obtain a deeper understanding of the properties of CSCs across five gastrointestinal cancers and to discuss their specific characteristics and markers. A better understanding of CSCs may ultimately contribute to improving patient prognoses and treatment outcomes and ultimately enhance the quality of life of gastrointestinal cancer sufferers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction to Gastrointestinal Cancers and Cancer Stem Cells
Gastrointestinal malignancies including oesophageal, gastric, colorectal, liver, and pancreatic cancers account for circa a quarter of all cancers; their prevalence is increasing and may have poor prognoses. Therapeutic efforts in these cancers include chemotherapy, surgery, and radiotherapy, and various predictive and prognostic biomarkers exist, but despite this, these cancers remain major health concerns and contribute to death due to cancer [1]. Irrespective of the type of cancer, a central premise of cancer biology attempts to explain the principles of cellular transformation and tumourigenesis and links these to the accumulation of genetic alterations that confer augmented fitness advantages to cells, and thereby, the cell pursues selfish positive selection programmes. Accordingly, non-mutually exclusive and potentially collaborative dichotomous stochastic and cancer stem cell (CSC) models have been proposed to explain this transformation [2,3,4,5]. To that end, the stochastic model attempts to explain within and between tumour heterogeneity and clonal evolution. The CSC model leans towards functional heterogeneity and states that not all cells within a tumour possess the same potential. Like normal tissue organisation, CSCs sit at the peak of their progeny’s hierarchical arrangement and will propagate the tumour. As mentioned, these models may be considered collaborative since certain clones may gain survival and growth advantages, become dominant within a tumour, and populate and propagate it [2,3,4,5,6].
CSCs may originate from tissue stem cells that undergone oncogenic transformation or from mutant progenitor cells that have acquired self-renewal capacity; hence, they will share key characteristics with stem cells such as heightened DNA repair, quiescence, and self-renewal capacities [5, 7, 8]. Accordingly, CSCs can self-renew to propagate the CSC population but can also undergo asymmetric division, resulting in a CSC and a more differentiated daughter cell [5]. CSCs may express efflux pumps, including ATP-binding cassettes that pump out compounds and toxins (e.g., Hoechst 33342 dye). CSCs also display aldehyde dehydrogenase 1 (ALDH1) activity that is linked to neutralising aldehydes and various metabolites, thereby reducing cellular damages due to reactive oxygen species (ROS) and consequently reducing levels of apoptosis [9,10,11,12,13]. In addition, the role of p53 in the genotoxic stress response mechanism has been studied, and the loss of p53 may also be relevant to CSC expansion, while in p53-incompetent cancer cells, EGFR may elevate ERK to increase sensitivity to drug treatment [14, 15]. Finally, CSCs account for tumour growth in immunodeficient animal models, including NOD-SCID gamma (NSG) mice [16]. Furthermore, CSCs can be impacted by and, in turn, influence the tumour microenvironment (TME) [17]. For example, CSCs in gastric cancer have been reported to reciprocally interact with cancer-associated fibroblasts and tumour-associated macrophages through various molecules such as TNF-α, MCP-1, TGF-β, and VEGFA [18]. Through their inherent properties and interactions with the TME, CSCs will therefore affect disease progression, metastasis, and relapse [12, 17, 18]. Despite these definitions and the extensively investigated properties and characteristics of CSCs, the CSC literature may interchangeably refer to terms such as cancer-initiating cells and CSCs. In the opinion of the author, however, cancer-initiating cells may encapsulate the cell of origin of cancer, while CSCs may indeed represent tumour-propagating cells and will constitute the cells intended in this review [19]. Accordingly, cancer-initiating cells, as the cells of origin, can be viewed as adult stem or progenitor cells within the tissue that obtain the first oncogenic event underlying transformation. For example, in colon cancer, krt19 + /Lrg − cells have been reported to represent cancer-initiating cells that can develop colonic adenomas, while at later stages of oncogenesis, CSCs will support tumour propagation and heterogeneity and, in the case of colon cancer will express markers such as CD144, CD44, and CD166. Even though both cell types possess self-renewal and multipotency capacities, they represent two distinct cell populations [20,21,22,23] (Fig. 1).
The process of tumourigenesis and the distinction between the cell of origin and CSCs. Various mutagenic and oncogenic molecules and processes leading to DNA damage and altered cellular and molecular processes will lead to the oncogenic transformation of the cell of origin of cancer. This cell will initiate the process of tumourigenesis, while a CSC will lead to the growth and propagation of the established cancer
CSCs may be detectable using in vitro and in vivo techniques, including cell sorting using various surface markers (e.g., CD44 and CD133), the efflux of Hoechst 33342 dye or other molecules, and sphere-and colony-formation assays. The use of 3D culture methods, including multicellular tumour spheroid and organoid formation, serves to bridge 2D culture and tumour formation in NSG mice and may be useful tools to test for CSCs [13, 16, 24,25,26]. Overall, the gold standard for assigning stemness to CSCs is their ability to propagate tumours in an NSG mouse model and thereby phenocopy the primary tumour, while serial transplantation using limiting dilutions may also indicate the occurrence and number of CSCs in tumours [5]. In this respect, CSCs are primarily responsible for driving and maintaining cancer states [27].
Given the attributed roles of CSCs, a better understanding of these cells in gastrointestinal cancers may contribute to lower tumour growth, metastasis, and recurrence [28]. This review will attempt to catalogue key markers and properties of CSCs in five gastrointestinal cancers.
CSCs in Gastrointestinal Cancers
Oesophageal Cancer
Oesophageal squamous cell carcinoma (ESCC) is the most common histological type of oesophageal cancer [29]. In this cancer, CSC-like characteristics can be triggered by the axis of MYC-miR-942-Wnt signalling pathways [30]. Furthermore, oesophageal squamous cell CSCs may be identified by markers inclusive of CD90, CD44, CD271, ALDH, Pygo2, Twist1, and MAML1, while they display the ability to form colonies in vitro in addition to tumourigenesis upon xenotransplantation to immunodeficient NOD/SCID mice and efflux of Hoechst 33342 dye [31, 32]. In addition, markers of these cells, such as ABCG2, SOX2, NANOG, ALDH1, and CD133, were linked to stemness and clinicopathological properties such as stage, recurrence, and prognosis [27]. For instance, the expression of ALDH1A1 (an isoform of ALDH) was associated with advanced stages of ESCC [33].
In a study, ESCC was shown to have elevated expression of the Wiskott-Aldrich syndrome protein and SCAR homologue (WASH) gene [34, 35]. WASH was involved in actin filament dynamics and linked to cancer stemness. CSCs were isolated from KYSE70 and KYSE450 cell lines by sphere formation assay. WASH was expressed in CSCs of ESCC, and its downregulation reduced the sphere-forming capacity of these cells by targeting IL-8 [34]. WASH downregulation led to reduced tumour growth in vivo upon xenografting to the immunodeficient mouse model (i.e., BALB/c nude mice), suggesting the WASH/IL-8 axis was linked to ESCC cell stemness [34].
In the more recent study by this group, the mechanistic link underlying the WASH/IL-8 regulatory axis was investigated [35]. Using bioinformatic analyses, the authors predicted that miR-637 targeted WASH. This was verified using KYSE70 and KYSE450 cell lines, in which miR-637 mimics downregulated WASH expression. In contrast, the miR-637 inhibitors in these cell lines led to increased WASH levels [35]. The authors also revealed direct binding of miR-637 to the 3′UTR of WASH by luciferase assay [35].
Additionally, miR-637 mimics and inhibitors could reduce and increase IL-8 levels, respectively, suggesting that IL-8 was also a target of miR-637. Furthermore, miR-637 mimics reduced sphere formation in vitro, while IL-8 overexpression could rescue the miR-637-mediated inhibition of the formation of spheres [35]. This link also applied to stemness factors SOX4, SOX9, NANOG, CD44, and ABCG2, which were reduced in cells treated with miR-367 mimics, while IL-8 overexpression rescued miR-367-mediated inhibition of these markers [35].
Furthermore, the authors attempted to establish the relationship between IL-8, WASH, and miR-637, whereby miR-637 inhibitors could increase IL-8 levels in controls but not in WASH knockdowns, revealing that miR-637 regulated IL-8 through targeting WASH. The same observation applied to sphere formation and the expression of stemness markers in that miR-637 inhibitors could increase sphere numbers and stemness markers in controls but not in WASH knockdowns. In association with earlier results, this suggested that miR-637 regulated cancer stemness through the WASH/IL-8 axis [35].
Finally, xenografting stably overexpressing miR-367 in the KYSE450 cell line into an immunodeficient mouse model led to attenuating tumour growth. In these in vivo samples, overexpressing miR-367 led to decreased WASH, IL-8, and stemness factors, confirming the in vitro data [35]. In conclusion, stemness markers and properties of oesophageal squamous cell CSCs may be regulated by miRNAs, cytoskeletal proteins, and cytokines (miR-637/WASH/IL-8), suggesting the effect of the TME on these cells as alluded to earlier [18] (Fig. 2).
CSCs in ESCCs. WASH is a protein involved in the cytoskeleton and is expressed in CSCs of ESCC. A WASH expression led to reduced IL-8 expression, colony formation, and tumour growth in vivo. B In KYSE70 and KYSE450 cell lines, miR-637 bound to the 3′-UTR of WASH and thereby targeted it. Inversely, the miR-637 inhibitor in these cell lines led to increased WASH. C miR-637 mimics could reduce IL-8 levels and, through that, sphere formation, while miR-637 inhibitors increased IL-8 levels. D The expression of stemness markers including SOX4, SOX9, NANOG, CD44, and ABCG2 was reduced in cells treated with miR-367 mimics, while IL-8 overexpression rescued this phenotype. E miR-637 inhibitors increased IL-8 levels in controls but not in WASH knockdowns. In addition, sphere formation and the expression of stemness markers such as SOX4, SOX9, NANOG, CD44, and ABCG2 were increased in cells treated with miR-637 inhibitors in controls but not in WASH knockdown cells. F Xenografting overexpressing miR-367 KYSE450 cell lines into a mouse model reduced tumour growth. In these in vivo samples, overexpressing miR-367 led to decreased WASH, IL-8, and stemness markers. KD, knockdown
Gastric Cancer
Gastric cancer (GC) is usually difficult to diagnose in its early stages, has a poor prognosis, and is a very common cancer [36]. The stemness characteristics of GC cells can also be highly dependent on signalling pathways such as the AKT-NF-κB-miR-21 axis [37]. A study reported that gastric cancer stem cells (GCSCs) cultured in suspension led to greater numbers of spheres and colonies compared to the GC line, SGC7901. In addition, they showed higher migration rates (tested by transwell assay) and expressed stemness markers such as CD24, CD44, SOX2, KLF4, and OCT4 [38,39,40].
With CD44/CD24 representing the high-confidence markers of GCSCs, a study expanded this marker list in primary tissue from 127 treatment-naive GC patients [41]. These samples were subjected to flow cytometric analyses, and putative GCSC markers inclusive of CD24, CD44, CD54, STRO-1, CD73, and EpCAM were tested [41]. The CD24 + CD44 + CD54 + EpCAM + subpopulation was then referred to as the gastric cancer cells with the extended phenotype (GCEPs) [41].
To test self-renewal and colony formation, AGS gastric cell lines were cultured in 3D conditions, and on the third day of cell culture, the tumourspheres displayed a GCEP content of circa 20%, while 80% of these GCEP cells expressed NANOG, OCT4, and SOX2. Interestingly, GCEP cells, in comparison to GCnEP cells (cells derived from the AGS tumourspheres with CD44 − CD24 − CD54 − EpCAM − surface marking), were challenged with secondary and tertiary tumoursphere formation capacity [41]. GCEPs but not GCnEPs formed secondary and tertiary tumourspheres providing more evidence of the CSC nature of the GCEP population [41].
These GCEPs were also detected in all GC patient tissue at a rate of 0.57–1.89% of the tumour population, and the authors set a cutoff of 1.19% to demarcate a low and high abundance of this population [41]. The authors found that a greater abundance of the CD24 + CD44 + CD54 + EpCAM + population correlated with higher TNM stage and cancer progression, providing evidence that the GCEPs were linked to disease stage and progression [41]. Despite this finding, the percentage of the GCEP population did not significantly impact the overall survival of patients [41].
The authors then applied this result to the zebrafish model to assay the tumourigenic potential of the GCEP compared to the GCnEP cell population. In evidence, AGS-derived tumourspheres were sorted to obtain GCEP and GCnEP cell populations, and these cells were then injected 48 h post-fertilisation into Tg(fli1:EGFP)y1 zebrafish larvae. The former group showed greater migration potential than the latter, induced angiogenesis, and migrated to the circulatory system [41]. GCnEPs showed the ability to promote tumour growth and disturb muscle structure, but their metastasis potential was much lower than GCEPs [41] (Fig. 3). In conclusion, the evidence supports the notion that the extended phenotype of CD24 + CD44 + CD54 + EpCAM + accurately represents the GCSCs.
CSCs in gastric cancer. A This study identified the GCEPs expressing CD24 + CD44 + CD54 + EpCAM + markers within the primary patient tissue for GC. AGS gastric cell lines were cultured in 3D conditions and formed tumourspheres. These tumourspheres displayed a 20% GCEP content, while of this 20% fraction, the majority expressed stemness markers such as NANOG, OCT4, and SOX2. AGS cell line derived GCEP cells (CD44 + CD24 + CD54 + EpCAM +) in comparison to GCnEPs cells (CD44 − CD24 − CD54 − EpCAM −), were challenged for self-renewal capacity. GCEPs, but not GCnEPs formed secondary and tertiary tumourspheres. B GCEP cells in GC patient samples varied from low to high (0.57–1.89%, respectively); the latter group (high) was linked to a higher TNM stage and progression. C The zebrafish model was used to assay the tumourigenic potential of the GCEP compared to the GCnEP cell population by injecting these cells into 48-h larvae. The former group showed greater migration potential than the latter, induced angiogenesis, and migrated to the circulatory system. GCnEPs showed the ability to promote tumour growth and disturb muscle structure, but their metastasis potential was much lower than that of GCEPs
Colorectal Cancer
Colorectal cancer is the second most common cause of patient death due to cancer. Colon cancer cells may be highly reliant on various signalling pathways, including the axes of resistin-TLR4-MyD88-ERK, in which resistin, a cytokine, could bind to TLR4 and ultimately lead to G1 phase arrest and reduced sensitivity to drug treatment [42]. From the viewpoint of colorectal CSCs, several groups outlined the key markers of these cells including CD133 + /CD44 + /ALDH1 + , EpCAM + /CD44 + , CD166 + , and CD44 + /CD24 + [23, 43, 44].
In a recent study by Nakano, colorectal CSCs were investigated using organoids (defined as miniature tissues that profile their complexity and heterogeneity) [45]. This group studied hyaluronan-mediated motility (RHAMM, also known as HMMR) expressed in a proportion of highly proliferative CD44 + human colorectal CSCs bearing self-renewal capacity using bulk and single-cell genomics. Initially, the transcriptomic profiles of CD44 + CSCs and CD44 − non-CSCs of colorectal cancer primary tissue and organoids were established [45]. The data was then subjected to GSEA gene ontology analysis, and it was shown that the genes linked to proliferation were expressed in the CD44 + cell populations obtained from both sources. Examples of these genes include PCNA, MKI67, TOP2A, survivin, and CDK4/6 [45]. Using the organoid models formed from patient tissue, the authors isolated 568 single cells and subjected them to transcriptomic analysis using SMART-seq. Principal component analysis and hierarchical clustering revealed the top 100 genes expressed by the two distinct subpopulations identified. For example, the subpopulation linked to high proliferation expressed genes such as MKI67, BIRC5, TOP2A, PCNA, CCNA2, FOXM1, and CDK1, while the subpopulation linked to low proliferation showed markers of differentiation such as CDX2 and KRT19 [45].
One gene that was of particular interest within the proliferative fraction was RHAMM, also known as HMMR, which correlated with genes linked to proliferation such as AURKA, MKI67, and FOXM1. It was observed that the CD44 population could be divided into RHAMM − CD44 + and RHAMM + CD44 + subpopulations. Interestingly, the latter subpopulations expressed genes such as MKI67, BIRC5, FOXM1, TOP2A, and PCNA, suggesting that this patient organoid-derived subpopulation is highly proliferative. The authors also found that primary tissue from colorectal cancer contains circa 25% RHAMM + CD44 + cells [45]. Further, transcriptomic analysis following the RHAMM + CD44 + cells showed the enrichment of proliferative genes such as MKI67, BIRC5, AURKA, TOP2A, and PCNA in this population compared to the RHAMM − CD44 + control consistent with previous results [45]. It was concluded that the RHAMM + CD44 + subpopulation was also associated with MAPK and SRC signalling pathways [45].
Furthermore, the authors tested the organoid-forming ability of the following four fractions: CD44 − RHAMM − , CD44 − RHAMM + , CD44 + RHAMM − , and CD44 + RHAMM + [45]. In evidence, the CD44 + fractions showed greater organoid-forming capacity compared to the CD44 − subpopulations (1.25% compared to zero), while within the CD44 + subpopulation, the CD44 + RHAMM + subpopulation formed more organoids than the CD44 + RHAMM − subpopulation (2% compared to 0.5%) [45], pointing towards heterogeneity within the CSC population that must be resolved on a clonal level [12].
Moreover, serial dilution coupled with xenografting to NSG mice revealed a higher CSC frequency in the CD44 + RHAMM + subpopulation, and this subpopulation also expressed CD166 and CD133 (CSC markers) [45]. Finally, the authors also showed that inhibiting RHAMM using shRNA knockdown suppressed organoid formation in vitro and tumour formation in vivo, suggesting that RHAMM maintained CSC capacities [45]. In conclusion, it is logical to state that RHAMM could be used as a marker for colorectal CSCs, was linked to high proliferative indices, and formed a bridge between CSCs and the TME (Fig. 4).
CSCs in colorectal cancer. A The transcriptional profile of CD44 + CSCs was established, and this data was subjected to GSEA gene ontology analysis, revealing associations between CD44 and proliferation markers such as PCNA, MKI67, TOP2A, survivin, and CDK4/6. B 568 cells of the patient-derived organoids (colorectal cancer) were subjected to single-cell analysis, principal component analysis (PCA), and hierarchical clustering. This analysis revealed populations with high or low proliferation rates expressing MKI67, BIRC5, TOP2A, PCNA, CCNA2, FOXM1, and CDK1 or CDX2 and KRT19, respectively. The authors then focused on the RHAMM gene within the proliferative fraction. The RHAMM + CD44 + subpopulation expressed genes such as MKI67, BIRC5, FOXM1, TOP2A, and PCNA. C In addition, the authors then tested this result in primary tissue from colorectal patients and found that circa 25% of this population is RHAMM + CD44 + and expressed genes like MKI67, BIRC5, AURKA, TOP2A, and PCNA. This population also showed enrichment for MAPK and SRC signalling pathways, CSC markers such as CD166 and CD133, and formed more organoids than their CD44 + RHAMM − counterparts. D The xenografting of organoid-derived cells after shRNA-mediated knockdown of RHAMM led to reduced tumour formation in vivo, while this knockdown suppressed organoid formation in vitro
Liver Cancer
Hepatocellular carcinoma is thought to be among the top 5 causes of death due to cancer [46]. Various factors may affect hepatocellular carcinoma. For example, HepG2-tumour-bearing mice fed glucose-containing water showed higher levels of PCSK9 and low-density lipoprotein cholesterol (LDLc). Mechanistically, PCSK9 increased glucose levels, ROS, and sterol regulatory element binding protein, while cells exposed to LDLc became desensitised to treatment [47].
From a CSC viewpoint, several markers of hepatocellular carcinoma, inclusive of CD133 + /CD49f + , CD90 + /CD45 − , CD13 + , and EpCAM + , have been suggested [48,49,50,51,52]. A recent study suggested that CD44v6 + cells represented the CSC population of hepatocellular carcinoma, and they attempted to understand the contribution of the HGF/Met signalling pathway to stemness properties in these cells [53]. In evidence, the authors attempted to investigate the tumourigenicity of CD44v6 + CSCs in vivo by isolating CD44v6 + and CD44v6 − from SNU398 liver cancer cell lines and xenografting them to BALB/c nude mice [53]. This resulted in CD44v6 + CSCs forming larger tumours and contributing to metastasis compared to their CD44v6 − counterparts. In addition, sections of the xenografted tumours were stained with a Ki67 proliferation marker, revealing the higher proliferative index of CD44v6 + CSCs [53].
Using in vitro assays also revealed that SNU398-derived CD44v6 + cells showed greater migration and invasion, colony- and sphere-formation, and proliferation capacities compared to the CD44v6 − controls [53]. Thus far, CD44v6 is a reliable marker of the liver CSC population, and the authors hypothesised that the HGF/MET pathway may be linked to the stemness of these cells. Firstly, it was shown that Met protein was highly expressed in hepatocellular cancer patient tumour tissue and was linked with shorter overall survival. Further, SNU398-derived CD44v6 + CSCs were shown to express stemness markers inclusive of Nanog, Oct4, and Sox2, and EMT-linked genes such as Snail1, Twist, and Slug compared to the CD44v6 − controls [53]. Furthermore, shRNA-mediated Met knockdown in the CD44v6 + subpopulation, followed by their xenografting to nude mice, led to smaller volumes of tumour growth in vivo, and this knockdown also led to reduced Nanog expression, suggesting links between Met and Nanog [53].
Furthermore, the levels of Met were manipulated using a Met inhibitor, PHA665752, in CD44v6 + and CD44v6 − cells, leading to the downregulation of stemness and EMT-linked genes [53]. Knocking down Met using lentiviral vectors also reduced sphere formation, and proliferation, in addition to reduced migration and invasion capacities, which could be rescued by Nanog overexpression [53]. shRNA-mediated Met knockdown led to reduced Nanog, ERK/p-ERK, and EMT-related genes, and these were rescued by overexpressing Nanog using LvNanog suggesting that Nanog is downstream of the HGF/MET/ERK pathway [53].
The xenograft model formed by orthotopically injecting CD44v6 + CSCs into the livers of these mice also showed Lvnanog could rescue Met knockdown-mediated reduced tumour volume [53]. In conclusion, CD44v6 + cells may represent CSCs in liver cancer, and these cells were linked to the HGF/MET/ERK pathway [53] (Fig. 5).
CSCs in liver cancer. A CD44v6 + cells presented in the CSC population can lead to greater Ki67 levels, tumour growth in vivo, and metastasis (BALB/c nude mice) compared to CD44v6 − cells. SNU398-derived CD44v6 + cells displayed greater migration, colony and sphere-forming, and proliferation capacities compared to controls. SNU398-derived CD44v6 + CSCs expressed stemness markers inclusive of Nanog, Oct4, and Sox2, and EMT-linked genes compared to the CD44v6 − controls. B shRNA-mediated Met knockdown in the CD44v6 + fraction, followed by their xenografting to the mouse model, led to smaller volumes of tumour growth and Nanog levels in vivo. C The levels of Met were manipulated using a Met inhibitor, PHA665752, in CD44v6 + and CD44v6 − cells leading to the downregulation of stemness and EMT-linked genes. Lentiviral knockdown of Met using lentiviral reduced sphere and colony formation in addition to reduced invasion capacities. D shRNA-mediated Met knockdown in SNU398 CD44v6 + led to reduced Nanog, ERK/p-ERK, and EMT-related genes and reduced migration, invasion, and proliferation capacities, all could be rescued by replacing Nanog (LvNanog). E The xenograft model formed by orthotopically injecting CD44v6 + CSCs into the livers of these mice also showed Lvnanog was able to rescue the Met shRNA phenotype
Pancreatic Cancer
Pancreatic cancer is among the top four leading causes of death due to cancer. Previous studies showed that PAF1, a component of the RNA polymerase II-associated factor 1 complex (PAF1C), could regulate CSCs in pancreatic cancer, and this can contribute to cancer progression [54]. From the viewpoint of CSCs, these cells in pancreatic cancer may express CD133 and CXCR4 [55]. In addition, Li and colleagues xenografted a subset of cells with high tumourigenic potential (pancreatic CSCs) bearing surface markers such as CD44/ CD24/ ESA (epithelial-specific antigen) to immunodeficient mice [56]. This subset of cells, representing less than 1% of pancreatic cancer cells, displayed a high tumourigenic potential [56]. Collectively, these two studies published in 2007 identified pancreatic cancer CSC [55, 56], while other groups also suggested other markers for this population may include Lrg5, Nanog, and DCLK1 [57, 58].
An interesting study by Ferrara and colleagues catalogued the stemness markers in cell lines and primary patient tissue of pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic cancer [59, 60]. This study selected 17 cell lines and 55 patient primary tissue of PDAC to study the expression of stemness markers and their links to in vitro and in vivo growth, tumourigenecity, and clonogenicity [60].
Initially, two main CSC subpopulations were identified: the ESA + CD24 + CD44 + fraction, ranging numerically from 0 to 99.35% within the population (median 3.46%), and the CD133 + CXCR4 + fraction, only representing a low proportion of circa 0–1.94% (median 0.13%) of the population, suggesting heterogeneity among and within the 17 PDAC cell lines [60]. The former surface markers were utilised to create a spectrum of stemness properties amongst the 17 PDAC cell lines. Accordingly, ESA + CD24 + CD44 + expression by these cell lines representing levels of stemness was arbitrarily classified as low, medium, and high, representing < 5%, 6–20%, and > 20% of the cell population, respectively [60]. On this scale, cell lines such as A8184, PaCa-44, and MIAPaCa-2 showed high, medium, and low percentages of cells within the population expressing ESA + CD24 + CD44 + markers, and this was assigned as high, medium, and low stemness potential, respectively [60]. The latter marker group (CD133 + CXCR4 +) was also segregated according to three levels of stemness, even though it usually represents a low percentage of the main population. In addition, the combination of CD24 + and/or CD133 + cells statistically correlated with the defined degrees of stemness (although ESA + , CD44 + , and CXCR4 + did not) [60].
Furthermore, the authors tested the population doubling time and cell cycle kinetics in the 17 PDAC cell lines and found that stemness (as defined by the percentage of cells expressing ESA + CD24 + CD44 +) was not associated with distribution in cell cycle phases (neither with the G1/G2 ratio nor the apoptosis rate). For example, MIAPaCa-2 had a higher proliferation rate than the A8184 cell line. This trend was also observed for clonogenic potential since stemness was not linked to clonogenic potential. For example, MIAPaCa-2 showed higher clonogenic potential than A8184 cells (despite the high stemness categorisation of the latter). Contrastingly, stemness was linked to tumour xenograft propagation time (in nude CD1 mice) since the low, medium, and high stemness groups showed in vivo tumour propagation times of 15, 5, and 4 days, respectively [60]. The study of the repertoire of their receptors and secreted molecules revealed that stemness levels in these cells were linked to KRT19, CA19.9, CD49a, IL-7, IL-10, CXCL9, and CCL3 proteins [60].
In addition, the 55 patient primary tissue samples were studied for various clinicopathological attributes, and it was revealed that CXCR4 + CD133 + and CD24 + cells were linked to mortality but ESA + CD24 + CD44 + were not. In conclusion, this study suggested heterogeneity within the CSC populations in PDAC and lower survival in patients expressing CD133, CD24, and CXCR4, and that some of the properties of PDAC cell lines are linked to their CSCs [60] (Fig. 6).
CSCs in PDAC cell lines. A The study outlines two main populations of cells with stemness properties: the ESA + CD24 + CD44 + fraction, ranging numerically from 0 to 99.35% within the population (median 3.46%), and the CXCR4 + CD133 + fraction, only representing a low fraction of circa 0–1.94% (median 0.13%) of the population. The percentage representation of the former group (ESA + CD24 + CD44 + fraction) was used to establish degrees of stemness in PDAC cell lines. For example, A8184, PaCa-44, and MIAPaCa-2 were assigned high, medium, and low stemness levels, respectively. B No correlation was found between the degree of stemness, proliferation and clonogenic potential; for example, A8184 showed a higher degree of stemness compared to MIAPaCa-2, while A8184 showed lower proliferation and clonogenic potential compared to MIAPaCA-2. C In contrast, stemness was inversely correlated with xenograft tumour development and propagation time. D The degree of stemness was correlated with secreted and non-secreted factors such as KRT19, CA19.9, CD49a, IL-7, IL-10, CXCL9, and CCL3
Overall, Table 1 summarises key aspects of the five cancer types reviewed in this study. The evidence to support new or more effective markers and regulatory networks governing stemness in these cancers was catalogued in this study (Table 2).
Discussion
In this study, the markers and properties of CSCs in five gastrointestinal cancers were reviewed.
Oesophageal cancer stem cells (ECSCs) were introduced as cells involved in the growth of tumours in vivo that may affect metastatic processes [61]. These CSCs may express CD90, CD44, CD271, ALDH, Pygo2, Twist1, and MAML1 [31, 32] and stemness markers such as ABCG2, SOX2, NANOG, ALDH1, and CD133 [27]. It was also revealed that miR-637 regulated sphere formation and cancer stemness through the WASH/IL-8 axis [34, 35]. In addition, miR-942 could increase tumour formation, CD90 population, and stemness markers in oesophageal cancer via the Wnt signalling pathway [30]. Consistent with the latter study, Cripto-1 (CR-1), a membrane protein, may also regulate stemness in ECSCs. Accordingly, CR-1 high and low cell populations were obtained from EC109 and TE-1 cell lines. The CR-1 high subpopulation showed higher expression of SOX2, OCT4, and NANOG compared to CR-1 low cells, and the knockdown of CR-1 led to reduce these proteins. Furthermore, self-renewal, tumourigenesis and in vitro and in vivo metastasis of the CR-1 high subpopulation were greater compared to the control [61], suggesting the importance of CR-1 in regulating stemness properties in oesophageal cancer, together with the previous studies outlining key features of these CSCs.
GCSCs may express markers such as CD24, CD44, SOX2, KLF4, and OCT4 [38,39,40]. A study showed that a subpopulation expressing CD24 + CD44 + CD54 + EpCAM + (GCEPs) represented the GCSCs, which formed secondary and tertiary tumourspheres and had greater migratory potential in vivo [41]. Consistently, GCSCs may express CD44 and its variants, and a study investigated a subpopulation (CD44V3 +) and revealed this variant was expressed in GC cell lines (e.g., MKN45) and patient-derived xenografts [62]. The CD44V3 + subpopulation also displayed CSC characteristics, including tumourigenic properties assayed using tumoursphere formation and in vivo tumour propagation in mouse models [62]. The latter involved limiting dilutions of panCD44 + /CD44V3– cells or CD44V3 + cells revealing their tumourigenic potential. Additionally, the CD44V3 + cells (akin to panCD44 + cells) developed distant metastases [62]. In addition, chromobox protein homolog 7 (CBX7), which is involved in GC proliferation and disease progression and is a member of the polycomb repressive complex, could regulate stemness (e.g., OCT4 and CD133 markers) through downregulating p16 and activating AKT and miR-21 [37]. Overall, these studies outline the properties of GCSCs.
Colorectal CSCs express CD133 + /CD44 + /ALDH1 + , EpCAM + /CD44 + , CD166 + , and CD44 + /CD24 + [23, 43, 44]. Also, a study proposed that RHAMM + CD44 + subpopulations may represent colorectal CSCs, and this population expressed genes such as MKI67, BIRC5, FOXM1, TOP2A, PCNA, CD166, and CD133, formed organoids, and led to tumour growth in vivo [45]. Consistently, the role of transformation/ transcription domain-associated protein (TRAPP), a regulator of NANOG, was investigated in regulating stemness in the colorectal cancer [63]. The study revealed direct binding between TRRAP and NANOG while overexpressing TRRAP blocked FBXW8-mediated proteasomal degradation of the NANOG protein [63]. Concerning the stemness-promoting properties of TRRAP in the HCT-15 colon cancer cell line, the authors showed that TRRAP downregulation reduced CD44 expression, tumour growth in the mouse xenograft model, and colony-forming capacity [63]. The latter could be rescued by overexpressing NANOG, revealing the TRRAP/NANOG axis in maintaining stemness properties [63], together outlining key features of these CSCs. Similarly, in colon cancer formation, the role of a signalling pathway inclusive of leptin-TNF-α-PUMA may be relevant since the knockdown of leptin led to TNF-α-mediated PUMA-executed apoptosis, outlining a fine balance between proliferation and cell death in this cancer [66].
Liver CSCs express CD133 + /CD49f + , CD90 + /CD45, CD13 + , and EpCAM + markers [48,49,50,51,52]. Furthermore, a study showed that CD44v6 + CSCs formed larger tumours in vivo and led to metastasis [53]. Additionally, CSCs in liver cancer may be regulated by non-coding RNA [64]. For example, deep sequencing revealed that in liver cancer primary tissue, miR‐HCC2 was upregulated, while in cell lines such as HepG2 and Huh7, miR-HCC2 was shown to promote stemness markers such as OCT4, SOX2, NANOG, and EpCAM, sphere formation, and CD90 + cell fraction [64]. In addition, using bioinformatic analyses and reporter assays, the authors revealed that this miRNA targeted BMP and activin membrane-bound inhibitor (BAMBI) to mediate Wnt signalling and thereby enhanced stemness properties in liver cancer cells. Accordingly, the YY1 transcription factor bound the miR-HCC2 promoter and activated it, thereby revealing the YY1/miR‐HCC2/BAMBI axis in promoting liver CSC stemness [64], collectively outlining the intricate network of factors regulating CSCs in this cancer. Interestingly, proliferation in hepatocellular carcinoma might be linked to the Wnt signalling pathway and glucose levels. Accordingly, high glucose levels triggered Wnt signalling via activating β-catenin, and suppressing DKK4, and this increased proliferation in culture. Also, in mice xenografted with HepG2 cells fed on high glucose diets, rapidly developed tumours with high levels of β-catenin, MYC, and reduced levels of DKK4 proliferation [67].
Pancreatic CSC markers may be CD133/ CXCR4 and CD44/ CD24/ ESA [55, 56, 60]. Other studies also defined markers of pancreatic CSCs as CD44, CD133, CD24, and ESA in addition to stemness markers such as Oct-4 and ALDH [65]. This study quantified the levels of expression of CD133, CD24, CD44, ESA, Nanog, Notch1, MDR1, and ABCG2 in CSCs compared to healthy pancreatic tissues. CSCs representing the CD133 + CD44 + CD24 + ESA + fraction highly expressed these genes [65]. This population was also subsequently xenografted to the NOD/SCID mouse model and formed tumours in vivo within 20 days compared to CD133 − CD44 − CD24 − ESA − cells which did not form tumours, outlining key features of these cells [65]. Interestingly, the role of PAF1C in regulating pancreatic CSCs was investigated, whereby PAF1 collaborated with DDX3 and PHF5A to regulate NANOG and other stemness genes in this cancer. The knockdown of PAF1 led to reduced tumour development in the orthotopic xenografted mouse model [54].
In conclusion, this study presented CSC markers and properties in five gastrointestinal cancers. Understanding CSC markers and processes will ultimately enhance our understanding of cancer and improve prognoses and treatment outcomes of patients.
Data Availability
Not applicable.
Abbreviations
- ALDH1:
-
Aldehyde dehydrogenase 1
- BAMBI:
-
BMP and activin membrane-bound inhibitor
- CR-1:
-
Cripto-1
- CBX7:
-
Chromobox protein homolog 7
- CSCs:
-
Cancer stem cells
- ESA:
-
Epithelial-specific antigen
- ESCC:
-
Oesophageal squamous cell carcinoma
- ECSC:
-
Oesophageal cancer stem cells
- GCEPs:
-
Gastric cancer cells with an extended phenotype
- GC:
-
Gastric cancer
- GCSC:
-
Gastric cancer stem cells
- KD:
-
Knockdown
- LDLc:
-
Low-density lipoprotein cholesterol
- NSG:
-
NOD-SCID gamma
- PDAC:
-
Pancreatic ductal adenocarcinoma
- PAF1C:
-
RNA polymerase II-associated factor 1 complex
- ROS:
-
Reactive oxygen species
- TME:
-
Tumour microenvironment
- TRAPP:
-
Transformation/transcription domain-associated protein
- WASH:
-
Wiskott-Aldrich syndrome protein and SCAR homologue
References
Kuntz S, Krieghoff-Henning E, Kather JN, Jutzi T, Höhn J, Kiehl L, et al. Gastrointestinal cancer classification and prognostication from histology using deep learning: Systematic review. Eur J Cancer. 2021;155:200–15.
Yap TA, Futreal PA, Pusztai L, Swanton C, Gerlinger M. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127ps10.
Fisher R, Swanton C, Pusztai L. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–85.
Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–44.
Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells–perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–44.
Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12(2):133–43.
Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. PNAS. 2003;100:3983–8.
Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13:727–38.
Cho RW, Clarke MF. Recent advances in cancer stem cells. Curr Opin Genet Dev. 2008;18(1):48–53.
Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–3.
Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68.
Akbari-Birgani S, Paranjothy T, Zuse A, Janikowski T, Cieślar-Pobuda A, Likus W, et al. Cancer stem cells, cancer-initiating cells and methods for their detection. Drug Discov Today. 2016;21(5):836–42.
Jerry DJ, Tao L, Yan H. Regulation of cancer stem cells by p53. Breast Cancer Res. 2008;10(4):304.
Kumari R, Chouhan S, Singh S, Chhipa RR, Ajay AK, Bhat MK. Constitutively activated ERK sensitizes cancer cells to doxorubicin: Involvement of p53-EGFR-ERK pathway. J Biosci. 2017;42(1):31–41.
Forde S, Matthews JD, Jahangiri L, Lee LC, Prokoph N, Malcolm TI, et al. Paediatric Burkitt lymphoma patient-derived xenografts capture disease characteristics over time and are a model for therapy. Br J Haematol. 2021;192(2):354–65.
Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem cell reports. 2014;2(1):78–91.
Ishimoto T, Sawayama H, Sugihara H, Baba H. Interaction between gastric cancer stem cells and the tumor microenvironment. J Gastroenterol. 2014;49(7):1111–20.
Jahangiri L, Ishola T, Pucci P, Trigg RM, Pereira J, Williams JA, et al. The role of autophagy and lncRNAs in the maintenance of cancer stem cells. Cancers. 2021;13(6):1239.
Chatterji P, Douchin J, Giroux V. Demystifying the differences between tumor-initiating cells and cancer stem cells in colon cancer. Curr Colorectal Cancer Rep. 2018;14(6):242–50.
Asfaha S, Hayakawa Y, Muley A, Stokes S, Graham TA, Ericksen RE, et al. Krt19(+)/Lgr5(−) cells are radioresistant cancer-initiating stem cells in the colon and intestine. Cell Stem Cell. 2015;16(6):627–38.
O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–10.
Dalerba P, Dylla SJ, Park I-K, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–63.
Sreepadmanabh M, Toley BJ. Investigations into the cancer stem cell niche using in-vitro 3-D tumor models and microfluidics. Biotechnol Adv. 2018;36(4):1094–110.
Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–84.
LaBarbera DV, Reid BG, Yoo BH. The multicellular tumor spheroid model for high-throughput cancer drug discovery. Expert Opin Drug Discov. 2012;7(9):819–83.
Islam F, Gopalan V, Lam AK. Detention and identification of cancer stem cells in esophageal squamous cell carcinoma. Methods Mol Biol. 2020;2129:177–91.
Taniguchi H, Moriya C, Igarashi H, Saitoh A, Yamamoto H, Adachi Y, et al. Cancer stem cells in human gastrointestinal cancer. Cancer Sci. 2016;107(11):1556–62.
Lam AKY. Molecular biology of esophageal squamous cell carcinoma. Crit Rev Oncol Hematol. 2000;33(2):71–90.
Ge C, Wu S, Wang W, Liu Z, Zhang J, Wang Z, et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget. 2015;6(13):10964–77.
Tang KH, Dai YD, Tong M, Chan YP, Kwanm PS, Fu L, et al. A CD90(+) tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res. 2013;73:2322–32.
Islam F, Gopalan V, Wahab R, Smith RA, Lam AK. Cancer stem cells in oesophageal squamous cell carcinoma: Identification, prognostic and treatment perspectives. Crit Rev Oncol Hematol. 2015;96(1):9–19.
Yang L, Ren Y, Yu X, Qian F, Bian B-S-J, Xiao H, et al. ALDH1A1 defines invasive cancer stem-like cells and predicts poor prognosis in patients with esophageal squamous cell carcinoma. Mod Pathol. 2014;27(5):775–83.
Huang L, Lian J, Chen X, Qin G, Zheng Y, Zhang Y. WASH overexpression enhances cancer stem cell properties and correlates with poor prognosis of esophageal carcinoma. Cancer Sci. 2017;108(12):2358–65.
Guo M, Lian J, Liu Y, Dong B, He Q, Zhao Q, et al. Loss of miR-637 promotes cancer cell stemness via WASH/IL-8 pathway and serves as a novel prognostic marker in esophageal squamous cell carcinoma. Biomark Res. 2022;10(1):77.
Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol. 2014;20(38):13842–62.
Ni S-J, Zhao L-Q, Wang X-F, Wu Z-H, Hua R-X, Wan C-H, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol. 2018;11(1):17.
Zhu Z, Xu J, Li L, Ye W, Xu G, Chen B, et al. Effect of gastric cancer stem cell on gastric cancer invasion, migration and angiogenesis. Int J Med Sci. 2020;17(13):2040–51.
Takaishi S, Okumura T, Tu S, Wang SSW, Shibata W, Vigneshwaran R, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27(5):1006–20.
Zhang C, Li C, He F, Cai Y, Yang H. Identification of CD44+CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol. 2011;137(11):1679–86.
Gómez-Gallegos AA, Ramírez-Vidal L, Becerril-Rico J, Pérez-Islas E, Hernandez-Peralta ZJ, Toledo-Guzmán ME, et al. CD24+CD44+CD54+EpCAM+ gastric cancer stem cells predict tumor progression and metastasis: clinical and experimental evidence. Stem Cell Res Ther. 2023;14(1):16.
Singh S, Chouhan S, Mohammad N, Bhat MK. Resistin causes G1 arrest in colon cancer cells through upregulation of SOCS3. Vol 591, FEBS letters. 2017. p 1371–82.
Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–5.
Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci USA. 2010;107(8):3722–7.
Nakano M, Taguchi R, Kikushige Y, Isobe T, Miyawaki K, Mizuno S, et al. RHAMM marks proliferative subpopulation of human colorectal cancer stem cells. Cancer Sci. 2023;00:1–12.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
Athavale D, Chouhan S, Pandey V, Mayengbam SS, Singh S, Bhat MK. Hepatocellular carcinoma-associated hypercholesterolemia: involvement of proprotein-convertase-subtilisin-kexin type-9 (PCSK9). Cancer Metab. 2018;6:16.
Rountree CB, Senadheera S, Mato JM, Crooks GM, Lu SC. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology. 2008;47(4):1288–97.
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13(2):153–66.
Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, Kim HM, et al. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120(9):3326–39.
Kimura O, Takahashi T, Ishii N, Inoue Y, Ueno Y, Kogure T, et al. Characterization of the epithelial cell adhesion molecule (EpCAM)+ cell population in hepatocellular carcinoma cell lines. Cancer Sci. 2010;101(10):2145–55.
Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123:1911–8.
Chen W, Wang R, Zhao Y, Li Y, Wang X, Peng W, et al. CD44v6+ hepatocellular carcinoma cells maintain stemness properties through Met/cJun/Nanog signaling. Stem Cells Int. 2022;2022:5853707.
Karmakar S, Rauth S, Nallasamy P, Perumal N, Nimmakayala RK, Leon F, et al. RNA polymerase II-associated factor 1 regulates stem cell features of pancreatic cancer cells, independently of the PAF1 complex, via interactions with PHF5A and DDX3. Gastroenterology. 2020;159(5):1898-1915.e6.
Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–23.
Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–7.
Amsterdam A, Raanan C, Schreiber L, Polin N, Givol D. LGR5 and Nanog identify stem cell signature of pancreas beta cells which initiate pancreatic cancer. Biochem Biophys Res Commun. 2013;433(2):157–62.
Ito H, Tanaka S, Akiyama Y, Shimada S, Adikrisna R, Matsumura S, et al. Dominant expression of DCLK1 in human pancreatic cancer stem cells accelerates tumor invasion and metastasis. PLoS ONE. 2016;11(1):e0146564.
Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008–20.
Ferrara B, Dugnani E, Sordi V, Pasquale V, Pellegrini S, Reni M, et al. A comprehensive characterization of stemness in cell lines and primary cells of pancreatic ductal adenocarcinoma. Int J Mol Sci. 2022;23(18):10663.
Liu Q, Cui X, Yu X, Bian B-S-J, Qian F, Hu X-G, et al. Cripto-1 acts as a functional marker of cancer stem-like cells and predicts prognosis of the patients in esophageal squamous cell carcinoma. Mol Cancer. 2017;16(1):81.
Giraud J, Seeneevassen L, Rousseau B, Bouriez D, Sifré E, Giese A, et al. CD44v3 is a marker of invasive cancer stem cells driving metastasis in gastric carcinoma. Gastric Cancer. 2023;26(2):234–49.
Kang K-T, Shin M-J, Moon H-J, Choi K-U, Suh D-S, Kim J-H. TRRAP enhances cancer stem cell characteristics by regulating NANOG protein stability in colon cancer cells. Int J Mol Sci. 2023;24(7):6260.
Gao H, Fan H, Xie H. The regulatory effect of the YY1/miR-HCC2/BAMBI axis on the stemness of liver cancer cells. Int J Oncol. 2023;62(5):59.
Shankar S, Nall D, Tang S-N, Meeker D, Passarini J, Sharma J, et al. Resveratrol inhibits pancreatic cancer stem cell characteristics in human and KrasG12D transgenic mice by inhibiting pluripotency maintaining factors and epithelial-mesenchymal transition. PLoS ONE. 2011;6(1):e16530.
Singh S, Mayengbam SS, Chouhan S, Deshmukh B, Ramteke P, Athavale D, et al. Role of TNFα and leptin signaling in colon cancer incidence and tumor growth under obese phenotype. Biochim Biophys Acta - Mol Basis Dis. 2020;1866(5):165660.
Chouhan S, Singh S, Athavale D, Ramteke P, Pandey V, Joseph J, et al. Glucose induced activation of canonical Wnt signaling pathway in hepatocellular carcinoma is regulated by DKK4. Sci Rep. 2016;6:27558.
Author information
Authors and Affiliations
Contributions
LJ wrote, reviewed, and edited this study.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jahangiri, L. Cancer Stem Cell Markers and Properties Across Gastrointestinal Cancers. Curr. Tissue Microenviron. Rep. 4, 77–89 (2023). https://doi.org/10.1007/s43152-023-00049-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43152-023-00049-8