Abstract
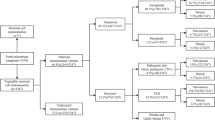
Chromosomal abnormality is one of the causes of fetal miscarriage. The potential differences of fetal chromosomal abnormalities in sporadic miscarriage (SM) and recurrent miscarriage (RM) remain unclear. The purpose of this study was to investigate copy number variations (CNVs) in SM and RM to provide useful genetic guidance for pregnancy and prenatal diagnosis. Four hundred eight samples of aborted fetuses were analyzed by CNV sequencing, and further functional enrichment analysis was performed. Chromosomal abnormalities were identified in 218 (53.4%) fetuses. There were 62 cases (15.2%) with structural chromosomal abnormalities, including 41 with VUS CNVs, 8 with pathogenic CNVs (pCNVs), and 5 with likely pCNVs. Duplications or deletions of 7p22, 8p22, 8p23, and Xp22.31 were significantly more common in RM cases and therefore believed to be related to RM. A total of 289 genes were identified, and 29 different functions were enriched as potential RM candidate genes and functions, which were mainly concentrated in 4 functional categories: chemokines and chemotaxis, protease activity and protein modification, defense response to bacterial and fungal infections, and immune response. The results of this study may improve our understanding of the etiology of RM and contribute to the establishment of a population-based genetic marker information for RM.



Similar content being viewed by others
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
La X, Wang W, Zhang M, Liang L. Definition and multiple factors of recurrent spontaneous abortion. Adv Exp Med Biol. 2021;1300:231–57.
Hu CY, Yang XJ, Hua XG, Jiang W, Huang K, Chen HB, Zhang XJ. Risk factors for spontaneous abortion from a prevention perspective in rural China: a population-based follow-up study. J Matern Fetal Neonatal Med. 2021;34:2583–91.
Moghbeli M. Genetics of recurrent pregnancy loss among Iranian population. Mol Genet Genomic Med. 2019;7:e891.
Definitions of infertility and recurrent pregnancy loss. a committee opinion. Fertil Steril. 2020;113:533–5.
Toth B, Würfel W, Bohlmann M, Zschocke J, Rudnik-Schöneborn S, Nawroth F, Schleußner E, Rogenhofer N, Wischmann T, von Wolff M, et al. Recurrent miscarriage: diagnostic and therapeutic procedures. Guideline of the DGGG, OEGGG and SGGG (S2k-Level, AWMF Registry Number 015/050). Geburtshilfe Frauenheilkd. 2018; 78:364–81
Department of Obstetrics, Chinese Society of Obstetrics and Gynecology. Chinese expert consensus on the diagnosis and tretment of recurrent spontaneous abortion. Chin J Obstet Gynecol. 2016;51:3–9.
Chan C, Ryu M, Zwingerman R. Preimplantation genetic testing for aneuploidy: a Canadian Fertility and Andrology Society Guideline. Reprod Biomed Online. 2021;42:105–16.
Khambata K, Raut S, Deshpande S, Mohan S, Sonawane S, Gaonkar R, Ansari Z, Datar M, Bansal V, Patil A, et al. DNA methylation defects in spermatozoa of male partners from couples experiencing recurrent pregnancy loss. Hum Reprod. 2021;36:48–60.
Bilal MY, Katara G, Dambaeva S. Clinical molecular genetics evaluation in women with reproductive failures. 2021; 85:e13313
Colley E, Hamilton S, Smith P, Morgan NV, Coomarasamy A, Allen S. Potential genetic causes of miscarriage in euploid pregnancies: a systematic review. Hum Reprod Update. 2019;25:452–72.
Ibrahim Y, Hotaling J. Sperm epigenetics and its impact on male fertility, pregnancy loss, and somatic health of future offsprings. Semin Reprod Med. 2018;36:233–9.
Wang Y, Li Y, Chen Y, Zhou R, Sang Z, Meng L, Tan J, Qiao F, Bao Q, Luo D, et al. Systematic analysis of copy-number variations associated with early pregnancy loss. Ultrasound Obstet Gynecol. 2020;55:96–104.
Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–83.
Lauer S, Gresham D. An evolving view of copy number variants. Curr Genet. 2019;65:1287–95.
Li Y, Anderson LA, Ginns EI, Devlin JJ. Cost effectiveness of karyotyping, chromosomal microarray analysis, and targeted next-generation sequencing of patients with unexplained global developmental delay or intellectual disability. Mol Diagn Ther. 2018;22:129–38.
Martinez-Portilla RJ, Pauta M. Added value of chromosomal microarray analysis over conventional karyotyping in stillbirth work-up: systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2019;53:590–7.
Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril. 2018;109:201–12.
Zhao X, Fu L. Efficacy of copy-number variation sequencing technology in prenatal diagnosis. J Perinat Med. 2019;47:651–5.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, Raca G, Ritter DI, South ST, Thorland EC, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22:245–57.
Brandt T, Sack LM. Adapting ACMG/AMP sequence variant classification guidelines for single-gene copy number variants. Genet Med. 2020;22:336–44.
Arredondo F, Noble LS. Endocrinology of recurrent pregnancy loss. Semin Reprod Med. 2006;24:33–9.
Venetis CA, Papadopoulos SP, Campo R, Gordts S, Tarlatzis BC, Grimbizis GF. Clinical implications of congenital uterine anomalies: a meta-analysis of comparative studies. Reprod Biomed Online. 2014;29:665–83.
Leisher SH, Balalian AA, Reinebrant H, Shiau S, Flenady V, Kuhn L, Morse SS. Systematic review: fetal death reporting and risk in Zika-affected pregnancies. Trop Med Int Health. 2021;26:133–45.
Muyayalo KP, Li ZH, Mor G. Modulatory effect of intravenous immunoglobulin on Th17/Treg cell balance in women with unexplained recurrent spontaneous abortion. Am J Reprod Immunol. 2018;80: e13018.
Lan L, She L, Zhang B, He Y, Zheng Z. Prenatal diagnosis of 913 fetuses samples using copy number variation sequencing. J Gene Med. 2021;23: e3324.
Zhang T, Sun Y, Chen Z, Li T. Traditional and molecular chromosomal abnormality analysis of products of conception in spontaneous and recurrent miscarriage. BJOG. 2018;125:414–20.
Shen J, Wu W, Gao C, Ochin H, Qu D, Xie J, Gao L, Zhou Y, Cui Y, Liu J. Chromosomal copy number analysis on chorionic villus samples from early spontaneous miscarriages by high throughput genetic technology. Mol Cytogenet. 2016;9:7.
Nagirnaja L, Palta P, Kasak L, Rull K, Christiansen OB, Nielsen HS, Steffensen R, Esko T, Remm M, Laan M. Structural genomic variation as risk factor for idiopathic recurrent miscarriage. Hum Mutat. 2014;35:972–82.
Dai R, Xi Q, Wang R, Zhang H, Jiang Y, Li L, Liu R. Chromosomal copy number variations in products of conception from spontaneous abortion by next-generation sequencing technology. Medicine (Baltimore). 2019;98: e18041.
Sheng YR, Hou SY, Hu WT, Wei CY, Liu YK, Liu YY, Jiang L, Xiang JJ, Sun XX, Lei CX, et al. Characterization of copy-number variations and possible candidate genes in recurrent pregnancy losses. Genes (Basel). 2021;12:141.
Li FX, Xie MJ, Qu SF, He D, Wu L, Liang ZK, Wu YS, Yang F, Yang XX. Detection of chromosomal abnormalities in spontaneous miscarriage by low-coverage next-generation sequencing. Mol Med Rep. 2020;22:1269–76.
Liu S, Song L, Cram DS, Xiong L, Wang K, Wu R, Liu J, Deng K, Jia B, Zhong M, Yang F. Traditional karyotyping vs copy number variation sequencing for detection of chromosomal abnormalities associated with spontaneous miscarriage. Ultrasound Obstet Gynecol. 2015;46:472–7.
Wang MZ, Lin FQ, Li M, He D, Yu QH, Yang XX, Wu YS. Semiconductor sequencing analysis of chromosomal copy number variations in spontaneous miscarriage. Med Sci Monit. 2017;23:5550–7.
Wu H, Huang Q, Zhang X, Yu Z, Zhong Z. Analysis of genomic copy number variation in miscarriages during early and middle pregnancy. Front Genet. 2021;12: 732419.
Luo S, Chen X, Yan T, Ya J, Xu Z, Cai P, Yuan D, Tang N. Application of copy number variation sequencing in genetic analysis of miscarriages in early and middle pregnancy. Cytogenet Genome Res. 2020;160:634–42.
Nagaoka K, Nojima H, Watanabe F, Chang KT, Christenson RK, Sakai S, Imakawa K. Regulation of blastocyst migration, apposition, and initial adhesion by a chemokine, interferon gamma-inducible protein 10 kDa (IP-10), during early gestation. J Biol Chem. 2003;278:29048–56.
Huang Y, Zhu XY, Du MR, Li DJ. Human trophoblasts recruited T lymphocytes and monocytes into decidua by secretion of chemokine CXCL16 and interaction with CXCR6 in the first-trimester pregnancy. J Immunol. 2008;180:2367–75.
Witkin SS, Linhares IM, Bongiovanni AM, Herway C, Skupski D. Unique alterations in infection-induced immune activation during pregnancy. BJOG. 2011;118:145–53.
Kwak-Kim J, Bao S, Lee SK, Kim JW, Gilman-Sachs A. Immunological modes of pregnancy loss: inflammation, immune effectors, and stress. Am J Reprod Immunol. 2014;72:129–40.
Tur-Torres MH, Garrido-Gimenez C, Alijotas-Reig J. Genetics of recurrent miscarriage and fetal loss. Best Pract Res Clin Obstet Gynaecol. 2017;42:11–25.
Szekeres-Bartho J, Šućurović S, Mulac-Jeričević B. The role of extracellular vesicles and PIBF in embryo-maternal immune-interactions. Front Immunol. 2018;9:2890.
D'Ippolito S, Tersigni C, Marana R, Di Nicuolo F, Gaglione R, Rossi ED, Castellani R, Scambia G, Di Simone N. Inflammosome in the human endometrium: further step in the evaluation of the "maternal side". Fertil Steril. 2016; 105:111–118.e111–114.
Banzai M, Sato S, Matsuda H, Kanasugi H. Trisomy 1 in a case of a missed abortion. J Hum Genet. 2004;49:396–7.
Vatin M, Burgio G, Renault G, Laissue P, Firlej V, Mondon F, Montagutelli X, Vaiman D, Serres C, Ziyyat A. Refined mapping of a quantitative trait locus on chromosome 1 responsible for mouse embryonic death. PLoS ONE. 2012;7: e43356.
Balogh A, Toth E, Romero R, Parej K, Csala D, Szenasi NL, Hajdu I, Juhasz K, Kovacs AF, Meiri H, et al. Placental galectins are key players in regulating the maternal adaptive immune response. Front Immunol. 2019;10:1240.
Mikwar M, MacFarlane AJ, Marchetti F. Mechanisms of oocyte aneuploidy associated with advanced maternal age. Mutat Res. 2020;785: 108320.
Xanthopoulou L, Ghevaria H, Mantzouratou A, Serhal P, Doshi A, Delhanty JD. Chromosome breakage in human preimplantation embryos from carriers of structural chromosomal abnormalities in relation to fragile sites, maternal age, and poor sperm factors. Cytogenet Genome Res. 2012;136:21–9.
Acknowledgements
The author would like to thank other colleagues who were not listed in the authorship of Department of Laboratory Medicine, Center for Precision Medicine and Center for Prenatal Diagnosis, Meizhou People’s Hospital (Huangtang Hospital), for their helpful comments on the manuscript.
Funding
This study was supported by the Guangdong Provincial Key Laboratory of Precision Medicine and Clinical Translation Research of Hakka Population under Grant 2018B030322003; the Science and Technology Program of Meizhou under Grant 2019B0202001; and the Key Scientific and Technological Project of Meizhou People’s Hospital under Grant MPHKSTP-20190102.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
This study was conducted on the basis of the Declaration of Helsinki and was supported by the Ethics Committee of the Meizhou People’s Hospital.
Consent to Participate
Each patient provided informed written consent, and the hospital’s ethics committees approved enrollment in the study.
Consent for Publication
Each participant had signed the consent form for the use of her personal data for research use and publication.
Conflict of Interest
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Zhang, X., Wu, H., Gu, Z. et al. Chromosomal Copy Number Variation Analysis in Pregnancy Products from Recurrent and Sporadic Miscarriage Using Next-Generation Sequencing. Reprod. Sci. 29, 2927–2936 (2022). https://doi.org/10.1007/s43032-022-00969-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-022-00969-0