Abstract
The timing of floral transition is critical for reproductive success in flowering plants. In long-day (LD) plant Arabidopsis, the floral regulator gene FLOWERING LOCUS T (FT) is a major component of the mobile florigen. FT expression is rhythmically activated by CONSTANS (CO), and specifically accumulated at dusk of LDs. However, the underlying mechanism of adequate regulation of FT transcription in response to day-length cues to warrant flowering time still remains to be investigated. Here, we identify a homolog of human protein arginine methyltransferases 6 (HsPRMT6) in Arabidopsis, and confirm AtPRMT6 physically interacts with three positive regulators of flowering Nuclear Factors YC3 (NF-YC3), NF-YC9, and NF-YB3. Further investigations find that AtPRMT6 and its encoding protein accumulate at dusk of LDs. PRMT6-mediated H3R2me2a modification enhances the promotion of NF-YCs on FT transcription in response to inductive LD signals. Moreover, AtPRMT6 and its homologues proteins AtPRMT4a and AtPRMT4b coordinately inhibit the expression of FLOWERING LOCUS C, a suppressor of FT. Taken together, our study reveals the role of arginine methylation in photoperiodic pathway and how the PRMT6-mediating H3R2me2a system interacts with NF-CO module to dynamically control FT expression and facilitate flowering time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Floral transition is controlled by several exogenous and endogenous developmental signals to ensure appropriate flowering time. To date, this transition is regulated by the photoperiodic, vernalization, autonomous, gibberellin, and the miR156-SPL module-mediated aging pathways (Wang et al. 2009; Andrés and Coupland 2012; He 2012; Bouché et al. 2017), in which the day length (photoperiod) is a stable seasonal signal to guarantee optimal flowering time in plants (Andrés and Coupland 2012; Romera-Branchat et al. 2014). Arabidopsis thaliana, a facultative long-day (LD) plant, can respond to photoperiodic signals to control floral transition by modulating the transcription of FLOWERING LOCUS T (FT) specifically in leaf vasculature via CONSTANS (CO)-dependent pathway (Andrés and Coupland 2012; Song et al. 2015). The B and C subunits of Nuclear Factor Y (NF-YB and NF-YC), as histone fold domain (HFD) NF-YB/NF-YC dimer, directly associate with CO thus compose of a trimeric NF-CO complex to regulate the FT expression (Wenkel et al. 2006; Gnesutta et al. 2018). Two NF-YB subunits (NF-YB2 and NF-YB3), and three NF-YC subunits (NF-YC3, NF-YC4, and NF-YC9) have been shown to interact with CO to activate FT expression (Kumimoto et al. 2010; Cao et al. 2014). Subsequently, FT protein, a major component of mobile florigen, transports from leaf veins to the shoot apex and leads to the floral transition (Andrés and Coupland 2012; Liu et al. 2013; Putterill and Varkonyi-Gasic 2016).
Except for the NF-CO complex, another regulator of FT expression is Polycomb group (PcG) which function to silence the transcription of FT (Wang et al. 2014; Müller-Xing et al. 2014; Luo et al. 2018). PcG proteins contain Polycomb repressive complex 1 (PRC1) and PRC2. PRC2 acts as a methyltransferase complex to catalyze histone 3 lysine-27 trimethylation (H3K27me3) on target chromatin, while PRC1 has been shown to maintain the H3K27me3 mark, leading to additional transcription repression by repressive chromatin modifications (Mozgova and Hennig 2015; Förderer et al. 2016). In Arabidopsis, two plant-unique BAH (Bromo adjacent homology) proteins SHORT LIFE (SHL) and EARLY BOLTING IN SHORT DAYS (EBS) form with a complex with EMBRYONIC FLOWER 1 (EMF1) to inhibit FT expression by reading the repressive H3K27me3 marks on FT locus (Li et al. 2018; Yang et al. 2018; Jing et al. 2019a). As another reader of H3K27me3, the LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) has been shown to maintain this mark on FT locus (Turck et al. 2007; Zhang et al. 2007), and LHP1 can also directly interact with EMF1 to silence FT expression and thus to repress photoperiodic flowering (Wang et al. 2014). Remarkably, the NF-CO module is partly co-activated to relieve Polycomb repression on the transcription of FT, resulting in the de-repression of FT that confer the LD induction of floral transition in Arabidopsis (Luo et al. 2018). In addition, other chromatin modifications such as histone deacetylation SIN3 LIKEs (SNLs) and chromatin remodeler PICKLE (PKL) are involved in the regulation of flowering by facilitating FT expression at dusk (Huang et al. 2019; Jing et al. 2019b). Although these studies have emphasized the significance of epigenetic regulation in photoperiodic flowering, it is still to be established whether and how other chromatin modifiers ‘communicate’ with NF-CO and NF-Y in response to inductive LDs to modulate FT expression.
Protein arginine methyltransferases (PRMTs) are responsible for catalyzing methylation at arginine (R) residues on histones that utilize S-adenosyl-L-methionine as a methyl donor. Besides, PRMTs-catalyzed non-histone substrates are involved in various biological processes including gene transcription, RNA processing and transport, cell signaling, DNA repair, and cell differentiation in mammals (Blanc and Richard 2017; Neault et al. 2012; Stein et al. 2012; Damez-Werno et al. 2016). In Arabidopsis, the posttranslational modifications of R mainly occur on histone 3 (H3) at R2, R8, R17, R26, and on H4 at only R3, and these modifications are catalyzed by different PRMTs (Ahmad and Cao 2012). PRMTs are divided into type I, comprising AtPRMT1a, AtPRMT1b, AtPRMT3, AtPRMT4a, AtPRMT4b, and AtPRMT6; type II, comprising AtPRMT5; type III, comprising only AtPRMT7; and a plant-specific AtPRMT10 (Ahmad and Cao 2012). These different types of PRMTs are responsible for producing symmetric ω-NG-monomethyl arginine (MMA), asymmetric ω-NG,NG-dimethylarginine (aDMA), and symmetric ω-NG,NG-dimethylarginine (sDMA) (Zurita-Lopez et al. 2012). Several studies indicate that PRMT4a/4b, PRMT5, and PRMT10 mediate histone arginine methylation to regulate flowering time by repressing the FLOWERING LOCUS C (FLC) expression (Niu et al. 2007, 2008; Pei et al. 2007; Schmitz et al. 2008). However, whether PRMTs involve in the regulation of photoperiodic FT expression remains unknown.
In this study, we identify a positive regulator of flowering PRMT6, which has been shown to catalyze the asymmetric dimethylation of R2 on H3 (H3R2me2a) (Guccione et al. 2007; Hyllus et al. 2007; Iberg et al. 2008). Further investigation finds that PRMT6 gene and its encoding protein accumulate in leaf veins at dusk, suggesting that PRMT6 may be associated with FT gene. PRMT6 interacts with NF-YCs and enhances their promotion on FT transcription. Moreover, AtPRMT6 and its homologues proteins AtPRMT4a and AtPRMT4b coordinately inhibit the expression of FLC, a suppressor of FT. Our results uncover the function of PRMT6 in plants, and provide insight into the role of arginine methylation in regulating photoperiodic flowering by ‘communicating’ with transcription factors.
Results
PRMT6 interacts with NF-Y subunits
Two subunits of NF-Y proteins (NF-YBs and NF-YCs) have been reported to interact with the CO and form NF-CO complex to regulate photoperiodic flowering (Hou et al. 2014; Gnesutta et al. 2018). We observed that two NF-YC members, NF-YC3 and its homolog NF-YC9, could interact with PROTEIN ARGININE METHYLTRANSFERASE 6 (AT3G20020; PRMT6) in yeast cells (Fig. 1A, B). Additionally, PRMT6 also strongly interacted with NF-YB3 in yeast cells, but did not interact with NF-YB2 and NF-YC4 (Fig. S1a). Next, we performed in vivo bimolecular fluorescence complementation (BiFC) assay to verify these interactions. The enhanced yellow fluorescent protein (EYFP) with non-fluorescent N-terminal was fused to the full-length CDS of NF-YC3, NF-YC9, and NF-YB3, and the C-terminal fragments were fused to PRMT6. When PRMT6-cEYFP and NF-YC3-nEYFP were co-expressed in Arabidopsis mesophyll protoplasts, the fluorescence was observed in nucleus, but not from protoplasts co-expressing NF-YC3-nEYFP and cEYFP alone, or PRMT6-cEYFP and nEYFP alone (Fig. 1C). Similarly, we also confirmed the interactions of PRMT6 with NF-YC9 (Fig. 1D), and PRMT6 with NF-YB3 (Fig. S1a, b). Concomitantly, a transient coimmunoprecipitation (Co-IP) assay was performed to testify the direct interaction of PRMT6 and NF-YC3 (Fig. S1c). We subsequently explored whether PRMT6 could physically associate with CO or FT protein, and found that PRMT6 did not interact with CO or FT in yeast cells (Fig. S1a).
Direct interactions of AT3G20020 (PRMT6) with NF-YC3 and NF-YC9 proteins. A, B Interactions of PRMT6 with NF-YC3 (A) and NF-YC9 (B) in yeast. Yeast cells harboring protein fusions with the DNA-binding domain (BD) and/or activation domain (AD) as indicated were grown on selective synthetic defined media lacking Trp, Leu, and His. C, D BiFC analysis of the interactions of PRMT6 with NF-YC3 (C) and NF-YC9 (D) in Arabidopsis protoplasts. Arabidopsis protoplasts were co-transformed transiently by a pair of plasmids. Yellowish-green signals indicate physical associations of paired proteins in the nuclei. Bar = 10 µm
PRMT6 enhances the promotion of NF-YC3;4;9 on flowering by facilitating FT expression
Analyses of the GUS reporter expression in transgenic plants expressing GUS under control of the PRMT6 promoter indicated that PRMT6 promoter was active in the vascular bundle cells (Fig. 2A). Subsequently, we measured the PRMT6 expression pattern every 4 h over a 24 h LD cycle in Col. PRMT6 mRNA abundance increased during daylight, peaks at ZT16 and then decreased (Fig. 2B). Additionally, we constructed a PRMT6-FLAG expression line driven by the native promoter region of PRMT6 and measured the protein abundance in the PRMT6-FLAG lines. PRMT6 protein varied diurnally and also peaked at ZT16 under LD conditions (Fig. 2C, D). Subcellular localization analysis showed that PRMT6 is a nuclear protein (Fig. S2). Collectively, nucleic protein PRMT6 displayed diurnal expression pattern and accumulated at dusk under LDs, which was consistent with the previously reported expression pattern of FT-GUS (Gu et al. 2013) and indicated that PRMT6 might modulate flowering by regulating FT transcription.
PRMT6 diurnally expressed at dusk in the vascular bundle cells under LDs. A Spatial expression patterns of PRMT6-GUS in 5-day-old seedlings, and 10-day-old seedlings of aerial part and cotyledon. Plants were stained for 6 h. Scale bars = 1 mm. B The relative transcription level of PRMT6 in 10-day-old Col seedlings under LDs. The transcription levels were normalized to UBQ10, and relative fold changes to Zeitgeber time 0 (ZT0) are presented. Bars indicate s.d. of triplicate measurements. White and dark bars below the x-axis indicate light and dark periods, respectively. C, D The expression levels of PRMT6-FLAG protein over a 24-h LD cycle examined by western blotting. Total proteins loaded in SDS-PAGE gels were stained with Coomassie Blue, antibody or the relative PRMT6-FLAG protein levels were normalized to H3 by the ImageJ program (D). The error bars indicate the s.d. measurements
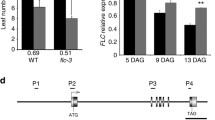
Next, two transfer DNA (T-DNA) insertion single-mutant prmt6-1 (Sail_385_A06) and prmt6-2 (Salk_151679C) (Figs. 3A; S3a) were used to explore its biological function in flowering. The total number of leaves of prmt6-1 and prmt6-2 mutant are not significantly different with Col under LD and SD conditions (Figs. 3B, C, S3b). To further confirm whether PRMT6 overexpression contributed to flowering time, we constructed overexpression lines and also found no significant differences between the Col and PRMT6 overexpression lines under LD conditions (Fig. S3c). We further explore whether PRMT6 mutation could affect the flowering phenotype of nf-yc3;4;9 mutants, and crossed prmt6-1 mutant with the nf-yc3;4;9 triple mutant to produce the quadruple mutant prmt6-1;nf-yc3;4;9. The total number of leaves in the prmt6-1;nf-yc3;4;9 was significantly more than that of nf-yc3;4;9 (Fig. 3B, D), suggesting that PRMT6 mutation could delay the flowering time of nf-yc3;4;9 mutant. Moreover, the expression of FT in prmt6-1;nf-yc3;4;9 mutant was lower than that of the nf-yc3;4;9 triple mutant line at ZT16 under LDs (Fig. 3E), in accordance with the additive action of prmt6-1 to delay flowering of nf-yc3;4;9.
Loss of PRMT6 function delays the floral transition of nf-yc3;4;9 by decreasing the FT expression under LDs. A Gene structure of PRMT6. Exons and 5’ untranslated region (UTR) or 3’ UTR are represented by black boxes and gray boxes, and arrows indicate transcription start sites (TSS); the T-DNA insertion sites of two lines are indicated with triangles. B Phenotype of Col, prmt6-1, prmt6-2, nf-yc3;4;9, and prmt6-1;nf-yc3;4;9 mutants grown in LDs. C Flowering times of the indicated genotypes grown in LDs. More than ten plants for each line were scored; bars indicated for standard deviation (s.d.); n.s. indicated non-significant difference. D Flowering times of the indicated lines grown in LDs. More than ten plants for each line were scored; bars indicated for s.d.; n.s. indicated non-significant difference; Double asterisks indicated statistically significant differences in the means between the indicated genotypes, as revealed by two-tailed Student’s t test (**p < 0.01). E Relative FT transcript levels in the seedlings of the indicated genotypes grown in LDs at ZT8 and ZT16. The transcript levels were first normalized to that of UBQ10. Bars indicate the s.d. of triplicate measurements
PRMT6 directly binds on the FT locus and affects its H3R2me2a levels
Because PRMT6 homologs have been identified to catalyze H3R2me2a in mammals (Guccione et al. 2007; Hyllus et al. 2007; Iberg et al. 2008), we performed sequence alignment of PRMT6 homologs in human, mouse, zebrafish, and Arabidopsis to test whether Arabidopsis PRMT6 have sequence similarity with PRMT6 homologs in animals. AtPRMT6 protein contained conserved domains including AdoMet methyltransferase (MTase) I, post-I, II, and III as well as the THW loop, which were involved in R methyltransferase activity (Fig. S4a, b).
We then examined whether the PRMT6 mutation could affect the global H3R2me2a levels. Histones were extracted from 10-day-old seedlings and probed with antibodies against H3R2me2a and H3. The global levels of H3R2me2a in prmt6-1 were similar to these in Col (Fig. S5a). Next, we performed chromatin immunoprecipitation (ChIP) assays to explore whether PRMT6 could bind on FT locus and affect its H3R2me2a methylation level. First, we constructed a PRMT6 antibody and confirmed its specificity (Fig. S5b). ChIP assays were conducted using 10-day-old prmt6-1 and Col seedlings grown under LDs. PRMT6 was enriched at the FT promoter region at ZT16 but not at ZT8 in Col compared to prmt6-1 mutant (Fig. 4A). Considering the NF-YCs’ transcript exhibits photoperiodic expression pattern under LD conditions (Fig. S6), we further performed ChIP experiments in Col, prmt6-1, nf-yc3;4;9, and prmt6-1;nf-yc3;4;9 lines at ZT8 and ZT16 of LDs, to verify whether PRMT6 could affect the H3R2me2a level to FT chromatin in the nf-yc3;4;9 mutant background. The H3R2me2a methylation level at FT locus of the prmt6-1; nf-yc3;4;9 quadruple mutant was significantly reduced at ZT16 but not at ZT8 (Fig. 4B). Collectively, these results demonstrated that PRMT6 mediated H3R2me2a modification to modulate FT transcription at the end of daylight.
PRMT6 modulates H3R2me2a methylation level and its binding on FT chromatin at dusk under LDs. A PRMT6 enrichment at the FT locus at midday (ZT8) and dusk (ZT16) under LDs. PRMT6 antibody was used to immunoprecipitate target chromatin extracted from Col or prmt6-1 (served as control). The amounts of immunoprecipitated genomic fragments were measured by RT-qPCR, and subsequently normalized to TUBLIN8 (TUB8). Error bars indicate s. d. from three technical replications. The regions examined in ChIP experiments are indicated with solid lines under FT structure. B H3R2me2a levels at the FT locus at dusk (ZT16) under LDs. The amounts of immunoprecipitated genomic fragments were quantified, and subsequently normalized to the input DNA. Error bars indicate s.d.
PRMT6 functions redundantly with PRMT4a and PRMT4b in the Arabidopsis genome
Previously, AtPRMT4a/4b (type I PRMT proteins) were shown to play important roles in regulating flowering time by influencing the transcription of FLC, which involved in vernalization and autonomous pathways (Niu et al. 2008). To clarify the genetic interaction between PRMT6 and PRMT4a/4b in regulating floral transition, we then generated prmt6-1;4a;4b and prmt6-2;4a;4b triple mutants by crossing. And the homozygous prmt6-1;4a;4b were used for subsequent analysis. The total leaf number of the prmt6-1;4a;4b and prmt6-2;4a;4b was more than prmt4a;4b double mutant, indicating that prmt6;4a;4b triple mutant exhibited the delayed flowering phenotype compared to prmt4a;4b double mutant under LD conditions (Fig. 5A–C). Next, we generated prmt6-1;4a;4b;ft-10 quadruple mutant lines to determine whether the ft-10 mutation could rescue the prmt6-1;4a;4b triple mutant phenotype. As expected, we found that the quadruple mutant line displayed a similar number of total leaves to the ft-10 single mutant (Fig. 5D), suggesting that the genetic mechanism regulating the prmt6-1;4a;4b phenotype could be completely rescued by the ft-10 mutation and that FT could thus be considered their downstream gene. Further investigation showed that the expression level of FLC in prmt6;4a;4b triple mutant was significantly higher than that in prmt4a;4b double mutant at ZT8 and ZT16 under LD conditions (Fig. 5E). PRMT6 mutation only inhibited the FT mRNA abundance in prmt6;4a;4b triple mutant at ZT16 under LD conditions (Fig. 5F). Together, these findings revealed that PRMT6 may be not only involved in regulation of photoperiodic FT expression through NF-CO module, but also exhibits redundancy with PRMT4a/PRMT4b to regulate FLC expression, thus to promote floral transition in Arabidopsis.
PRMT6 mutation enhances the late flowering phenotype of prmt4a;4b double mutant. A Phenotypes of Col, prmt6-1, prmt4a;4b, and prmt6-1;4a;4b mutants. Plants were grown under LD conditions. The white arrow indicates that the prmt4a;4b double mutants were bolting. B Flowering time of Col, prmt6-1, prmt4a;4b, and prmt6-1;4a;4b under LD conditions. More than 15 plants for each line were scored; bars indicate the s.d.; significant differences between the means of the indicated genotypes were revealed by a two-tailed Student’s t test (n.s., no significant difference; *p < 0.05; **p < 0.01). C Flowering time of Col, prmt6-2, prmt4a;4b, and prmt6-2;4a;4b grown under LD conditions. D Flowering time of Col, prmt6-1;4a;4b, ft-10, and prmt6-1;4a;4b ft-10 grown under LD conditions. E, F the relative transcription levels of FLC (E) and FT (F) in seedlings of the indicated genotypes as quantified by RT-qPCR. The transcription levels were normalized to UBQ10, and the fold changes relative to Col are presented
Discussion
PRMT6 serves as co-factor of NF-YCs and PRMT4s during flowering
In animal systems, PRMT6 has been reported to play an important role in the regulation of disease and developmental processes (Boulanger et al. 2005; Yang and Bedford 2013), and acts as writers to catalyze H3R2me2a (Guccione et al. 2007; Hyllus et al. 2007; Iberg et al. 2008). However, the function of PRMT6 of regulating plant development remains unclear. In this study, we have first illustrated the involvement of AtPRMT6, an Arabidopsis homolog of human HsPRMT6, in the flowering transition. In Arabidopsis prmt6 mutants exhibited a similar phenotype to wild type, whereas PRMT6 mutation further delays the flowering time of prmt4a;4b (Fig. 5; Niu et al. 2008), suggesting PRMT6 could function redundantly with PRMT4a/PRMT4b in controlling floral transition. In addition, our results indicated that the PRMT6 mutation also strengthened the late flowering phenotype of nf-yc3;4;9 (Fig. 3). These results demonstrated PRMT6, which might serve as a co-factor of NF-YCs and PRMT4s, synergistically modulated floral transition.
PRMT6 control floral transition via NF-Y-CO module or FLC-dependent pathway
In Arabidopsis, movement of the FT protein (also known as florigen) contributes to inducing the flowering transition through long-distance signaling from the leaf vascular tissue (phloem) to the SAM (Corbesier et al. 2007), and FT-GUS was mainly expressed in the leaf vascular tissues (Gu et al. 2013). Our study found that PRMT6 exhibited a similar expression pattern to that of FT in the leaf phloem (Fig. 2A, B). Moreover, the expression patterns of Flag-PRMT6 also showed rhythmic oscillations under LD conditions and accumulated at dusk under LDs (Fig. 2C), coinciding with the peak in CO protein level and increasing FT expression. These results demonstrated that PRMT6 could regulate CO-FT module. The nuclear factor NF-Y could bind to target DNA sequences accumulated on silent chromatin regions and act as a ‘pioneer’ to open up the chromatin structure to activate gene expression (Fleming et al. 2013; Oldfield et al. 2014). In addition, its subunits have been reported to interact with the CO protein (forming NF-CO complexes) to regulate flowering time (Gnesutta et al. 2018). In our study, PRMT6 was associated with NF-YC proteins, but did not interact with CO in yeast cells (Fig. S1). Considering the interaction between NF-YC proteins with CO testified by the previous studies (Hou et al. 2014; Gnesutta et al. 2018), we suppose that PRMT6 could interplay with CO by NF-YC proteins. Moreover, we confirmed the occupancy of PRMT6 at FT loci, as well as the reduced H3R2me2a modification in prmt6-1;nf-yc3;4;9 compared with nf-yc3;4;9 (Fig. 4). These data demonstrated that NF-YC proteins could recruit PRMT6 to FT promoter, consequently catalyzing H3R2me2a modification and finally accelerate floral transition. On the other hand, we observed PRMT6 and PRMT4 proteins synergistically inhibited the expression of FLC (Fig. 5). Taken together, PRMT6, NF-YCs, and PRMT4s synergistically modulated floral transition by CO-FT module or FLC-related pathway.
In conclusion, our study revealed that AtPRMT6, a PRMT6 homolog in Arabidopsis, acts as a positive regulator of floral transition. AtPRMT6 physically associates with three NF-Y subunits to bind to the FT locus around dusk (ZT16) of LDs, consequently changing the methylation abundance on FT locus to promote its expression at ZT16 of LDs. In addition, PRMT6 play a redundant role with PRMT4a/PRMT4b via inhibiting FLC expression during floral transition (Fig. 6). Our study reveals the role of arginine methylation in photoperiodic pathway and how the PRMT6-mediating H3R2me2a system interacts with NF-CO module to dynamically control FT expression and facilitate flowering time.
A working model for PRMT6 promoting flowering time in Arabidopsis. AtPRMT6 physically associates with three NF-Y subunits to bind to the FT locus around dusk (ZT16) of LDs, consequently changing the methylation abundance on FT locus to promote its expression at ZT16 of LDs. In addition, PRMT6 play a redundant role with PRMT4a/PRMT4b via inhibiting FLC expression during floral transition
Materials and methods
Plant materials, growth conditions, and flowering time
The ft-10 (Kardailsky et al. 1999), prmt4a;4b (Niu et al. 2008), and nf-yc3;4;9 (Hou et al. 2014) mutants were described previously. Two prmt6 lines, prmt6-1 (Sail_385_A06) and prmt6-2 (Salk_151679C), were obtained from the ABRC. Plants were grown in LD conditions (16-h light/8-h dark) or SD conditions (8-h light/16-h dark) under cool white fluorescent light at 22 °C. Total leaf numbers (rosette and cauline leaves) were calculated as a measurement of flowering time. Approximately 15 plants were analyzed for each line.
RNA extraction and RT-qPCR
Total RNA was extracted from the aerial parts of 10-day-old seedlings grown under LD conditions using the Qiagen RNeasy Plus Mini Kit and then reverse-transcribed into cDNA with M-MLV reverse transcriptase and oligo (dT) primers (Promega). Real-time quantitative PCR (RT-qPCR) was performed using the Roche Light Cycler 480 II System using SYBR Green PCR master mix, as described previously (Gu et al. 2013). Each sample was quantified at least three times and normalized to UBQ10. The relative transcription levels was calculated as 2−△Ct (Gu et al. 2013). The primer pairs for FT, NF-YC3, NF-YC9, TUB2, and UBQ10 amplification were described previously (Gu et al. 2013; Hou et al. 2014).
Plasmid construction and plant transformation
To generate the pPRMT6-PRMT6:FLAG plasmid, the full-length PRMT6 CDS without the stop codon was first fused with a 3 × FLAG tag and cloned into pHGW vector via Gateway technology. The sequences of the primers used for plasmid construction are specified in Table S1. Agrobacterium tumefaciens GV3101 was used to generate stable transgenic lines.
Histochemical β-glucuronidase staining
To construct AtPRMT6-GUS, 4.5 kb genomic fragments including the 2.4 kb promoter and 2.1 kb genomic coding sequence of PRMT6 (including exons and introns) were inserted into the pMDC162 vector via Gateway technology (Invitrogen). Histochemical β-glucuronidase (GUS) staining was performed as described previously (Gu et al. 2013). Briefly, GUS staining was carried out with 5- and 10-day-old seedlings after hygromycin selection by immersing them in X-Gluc (5-bromo-4-chloro-3-indoyl β-d-glucuronide) (0.5 mg/ml) (Gold Biotechnology; USA), followed by vacuum briefly to ensure uniform staining. The tissues were then incubated in X-Gluc at 37 °C for various times followed by incubation in 70% ethanol at 37 °C overnight to remove the chlorophyll from the green tissues.
Subcellular localization
For PRMT6–GFP fusion protein construction, the 1.3 kb CDS (coding sequence) region containing all the exons of PRMT6 was inserted between the 35S promoter and GFP in the pMDC85-GFP vector via Gateway technology (Invitrogen). The subcellular localization of PRMT6 was conducted as described previously (Gu et al. 2013). GFP fluorescence signals were observed and recorded using a Zeiss LSM 700 confocal laser scanning microscope.
Preparation of polyclonal antibody
The synthetic PRMT antibody was performed as described previously (Geng et al. 2020; Zhang et al. 2021). Briefly, a 15-amino acid fragment of PRMT6 (residues 105–119; TYREAIMQHQSLIEG) was synthesized by the solid-phase peptide synthesis (SPPS) method and confirmed by mass spectrometry (MS) and high-performance liquid chromatography (HPLC). The polypeptide was coupled through the Sulfo-SMCC agent to react with keyhole limpet hemocyanin (KLH) for preparation of the immune complex. The prepared immune complex was immunized to two rabbits to produce the antibody and then purified by a specific affinity chromatography column. The synthesized antibody was verified by western blot analysis using total proteins extracted from wild-type and prmt6-1 plants.
Histone extraction and immunoblotting
Histone protein extraction and western analysis were performed as described previously (Zhang et al. 2021). Briefly, total histones were extracted from 10-day-old seedlings grown in LD conditions, separated on an SDS-PAGE gel, and subsequently transferred to a 0.2-μm nitrocellulose membrane (Bio-Rad). The protein blots were probed with anti-H3 (Abcam, Cat#: ab1791) and H3R2me2a (Abcam, Cat#: ab175007). Immunoblotting was visualized by chemiluminescence. Blotting signals were captured using ImageJ software, and the relative protein level of H3R2me2a was normalized to that of H3. Experiments were repeated at least two biological times.
Yeast two-hybrid (Y2H) assay
Yeast two-hybrid assays were conducted using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech) as per the manufacturer’s instructions. The full-length coding sequences of PRMT6, NF-YC3, NF-YC4, NF-YC9, NF-YB3, CO, and FT were cloned into the pGADT7 and pGBKT7 vectors and subsequently introduced into the yeast strain AH109. Yeast cells were spotted on selective media lacking leucine (L), tryptophan (W), histidine (H), and adenine (A) for interaction detection or on drop-out media lacking L and W (as control).
Bimolecular fluorescence complementation (BiFC) assay
The full-length coding sequences for PRMT6, NF-YC3, NF-YC9, and NF-YB3 were fused with the coding sequence for an N-terminal EYFP fragment in the nEYFP-N1/pUGW0 (P35S/N-nEYFP) vector and/or for a C-terminal EYFP fragment in the cEYFP-N1/pUGW0 (P35S/N-cEYFP) vector (Nakagawa et al. 2007). Plasmid pairs were assessed by a transient expression system using Arabidopsis mesophyll protoplast (Yoo et al. 2007). Within 12–18 h, the EYFP fluorescence emitted from the Arabidopsis mesophyll protoplast was imaged with a Leica TCS SP8 laser scanning confocal microscope (Leica).
Co-immunoprecipitation (Co-IP) assay
Co-immunoprecipitation (Co-IP) experiments were carried out as previously described with some modifications (2013). Briefly, plasmid pairs were performed by a transient expression system using Arabidopsis mesophyll protoplast (Yoo et al. 2007). After 12–18 h incubation, total proteins were extracted from Arabidopsis mesophyll protoplasts and immunoprecipitated with anti-FLAG M2 affinity gel (Sigma, Cat#: A2220), and the immunoprecipitated protein was detected by western blotting with anti-FLAG (Sigma, Cat#: A8592).
Chromatin immunoprecipitation (ChIP) and ChIP-qPCR analysis
ChIP experiments were carried out as previously described (Gu et al. 2013). Briefly, total chromatin was extracted from 10-day-old seedlings grown under LD conditions and immunoprecipitated with anti-H3R2me2a (Abcam, Cat#: ab175007) and anti-PRMT6 (synthesized in this study). Quantitative PCR (qPCR) was conducted to measure the amounts of FT and the constitutively expressed TUB8 fragments on a Roche Light Cycler 480 II System using SYBR Green PCR master mix. The ChIP-qPCR primer pairs for FT, were described previously (Gu et al. 2013).
Accession numbers
Sequence data from this paper can be found in The Arabidopsis Information Resource (TAIR) website (http://www.arabidopsis.org/) under the following accession numbers: PRMT6, At3g20020; CO, At5g15840; FT, At1g65480; NF-YC3, At1g54830; NF-YC4, At5g63470; NF-YC9, At1g08970; NF-YB2, At5g47640; NF-YB3, At4g14540.
Data availability
Not applicable.
Code availability
Not applicable.
Change history
24 December 2021
A Correction to this paper has been published: https://doi.org/10.1007/s42994-021-00066-x
References
Ahmad A, Cao X (2012) Plant PRMTs broaden the scope of arginine methylation. J Genet Genom 39:195–208. https://doi.org/10.1016/j.jgg.2012.04.001
Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13:627–639. https://doi.org/10.1038/nrg3291
Blanc RS, Richard S (2017) Arginine methylation: the coming of age. Mol Cell 65(1):8–24. https://doi.org/10.1016/j.molcel.2016.11.003
Bouché F, Woods DP, Amasino RM (2017) Winter Memory throughout the plant kingdom: different paths to flowering. Plant Physiol 173:27–35. https://doi.org/10.1104/pp.16.01322
Boulanger M-C, Liang C, Russell RS et al (2005) Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol 79:124–131. https://doi.org/10.1128/JVI.79.1.124-131.2005
Cao S, Kumimoto RW, Gnesutta N et al (2014) A distal CCAAT/nuclear factor Y complex promotes chromatin looping at the flowering locus T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26:1009–1017. https://doi.org/10.1105/tpc.113.120352
Corbesier L, Vincent C, Jang S et al (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033. https://doi.org/10.1126/science.1141752
Damez-Werno DM, Sun H, Scobie KN et al (2016) Histone arginine methylation in cocaine action in the nucleus accumbens. Proc Natl Acad Sci 113:9623–9628. https://doi.org/10.1073/pnas.1605045113
Fleming JD, Pavesi G, Benatti P et al (2013) NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res 23:1195–1209. https://doi.org/10.1101/gr.148080.112
Förderer A, Zhou Y, Turck F (2016) The age of multiplexity: recruitment and interactions of polycomb complexes in plants. Curr Opin Plant Biol 29:169–178. https://doi.org/10.1016/j.pbi.2015.11.010
Geng Y, Zhang P, Liu Q et al (2020) Rice homolog of Sin3-associated polypeptide 30, OsSFL1, mediates histone deacetylation to regulate flowering time during short days. Plant Biotechnol J 18:325–327. https://doi.org/10.1111/pbi.13235
Gnesutta N, Mantovani R, Fornara F (2018) Plant flowering: imposing DNA specificity on histone-fold subunits. Trends Plant Sci 23:293–301. https://doi.org/10.1016/j.tplants.2017.12.005
Gu X, Wang Y, He Y (2013) Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol 11:e1001649. https://doi.org/10.1371/journal.pbio.1001649
Guccione E, Bassi C, Casadio F et al (2007) Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449:933–937. https://doi.org/10.1038/nature06166
He Y (2012) Chromatin regulation of flowering. Trends Plant Sci 17:556–562. https://doi.org/10.1016/j.tplants.2012.05.001
Hou X, Zhou J, Liu C et al (2014) Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun 5:4601. https://doi.org/10.1038/ncomms5601
Huang F, Yuan W, Tian S et al (2019) SIN3 LIKE genes mediate long-day induction of flowering but inhibit the floral transition in short days through histone deacetylation in Arabidopsis. Plant J 100:101–113. https://doi.org/10.1111/tpj.14430
Hyllus D, Stein C, Schnabel K et al (2007) PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev 21:3369–3380. https://doi.org/10.1101/gad.447007
Iberg AN, Espejo A, Cheng D et al (2008) Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem 283:3006–3010. https://doi.org/10.1074/jbc.C700192200
Jing Y, Guo Q, Lin R (2019a) The B3-domain transcription factor VAL1 regulates the floral transition by repressing flowering locus T. Plant Physiol 181:236–248. https://doi.org/10.1104/pp.19.00642
Jing Y, Guo Q, Lin R (2019b) The chromatin-remodeling factor pickle antagonizes polycomb repression of FT to promote flowering. Plant Physiol 181:656–668. https://doi.org/10.1104/pp.19.00596
Kardailsky I, Shukla VK, Ahn JH et al (1999) Activation tagging of the floral inducer FT. Science 286:1962–1965. https://doi.org/10.1126/science.286.5446.1962
Kumimoto RW, Zhang Y, Siefers N, Holt BF (2010) NF-YC3, NF-YC4 and NF-YC9 are required for constans-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J 63:379–391. https://doi.org/10.1111/j.1365-313X.2010.04247.x
Li Z, Fu X, Wang Y et al (2018) Polycomb-mediated gene silencing by the BAH–EMF1 complex in plants. Nat Genet 50:1254–1261. https://doi.org/10.1038/s41588-018-0190-0
Liu L, Zhu Y, Shen L, Yu H (2013) Emerging insights into florigen transport. Curr Opin Plant Biol 16:607–613. https://doi.org/10.1016/j.pbi.2013.06.001
Luo X, Gao Z, Wang Y et al (2018) The nuclear factor-constans complex antagonizes Polycomb repression to de-repress flowering locus T expression in response to inductive long days in Arabidopsis. Plant J 95:17–29. https://doi.org/10.1111/tpj.13926
Mozgova I, Hennig L (2015) The Polycomb group protein regulatory network. Annu Rev Plant Biol 66:269–296. https://doi.org/10.1146/annurev-arplant-043014-115627
Müller-Xing R, Clarenz O, Pokorny L et al (2014) Polycomb-group proteins and FLOWERING LOCUS T maintain commitment to flowering in Arabidopsis thaliana. Plant Cell 26(6):2457–2471. https://doi.org/10.1105/tpc.114.123323
Nakagawa T, Kurose T, Hino T et al (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41. https://doi.org/10.1263/jbb.104.34
Neault M, Mallette FA, Vogel G et al (2012) Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucl Acids Res 40:9513–9521. https://doi.org/10.1093/nar/gks764
Niu L, Lu F, Pei Y et al (2007) Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep 8:1190–1195. https://doi.org/10.1038/sj.embor.7401111
Niu L, Zhang Y, Pei Y et al (2008) Redundant requirement for a pair of protein arginine methyltransferase4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol 148:490–503. https://doi.org/10.1104/pp.108.124727
Oldfield AJ, Yang P, Conway AE et al (2014) Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol Cell 55:708–722. https://doi.org/10.1016/j.molcel.2014.07.005
Pei Y, Niu L, Lu F et al (2007) Mutations in the type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol 144:1913–1923. https://doi.org/10.1104/pp.107.099531
Putterill J, Varkonyi-Gasic E (2016) FT and florigen long-distance flowering control in plants. Curr Opin Plant Biol 33:77–82. https://doi.org/10.1016/j.pbi.2016.06.008
Romera-Branchat M, Andrés F, Coupland G (2014) Flowering responses to seasonal cues: what’s new? Curr Opin Plant Biol 21:120–127. https://doi.org/10.1016/j.pbi.2014.07.006
Schmitz RJ, Sung S, Amasino RM (2008) Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci U S A 105:411–416. https://doi.org/10.1073/pnas.0710423104
Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66:441–464. https://doi.org/10.1146/annurev-arplant-043014-115555
Stein C, Riedl S, Rüthnick D et al (2012) The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucl Acids Res 40:9522–9533. https://doi.org/10.1093/nar/gks767
Turck F, Roudier F, Farrona S et al (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3:e86. https://doi.org/10.1371/journal.pgen.0030086
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749. https://doi.org/10.1016/j.cell.2009.06.014
Wang Y, Gu X, Yuan W et al (2014) Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev Cell 28:727–736. https://doi.org/10.1016/j.devcel.2014.01.029
Wenkel S, Turck F, Singer K et al (2006) Constans and the CCAAT box binding complex Share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984. https://doi.org/10.1105/tpc.106.043299
Yang Y, Bedford MT (2013) Protein arginine methyltransferases and cancer. Nat Rev Cancer 13:37–50. https://doi.org/10.1038/nrc3409
Yang Z, Qian S, Scheid RN et al (2018) EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat Genet 50:1247–1253. https://doi.org/10.1038/s41588-018-0187-8
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572. https://doi.org/10.1038/nprot.2007.199
Zhang X, Germann S, Blus BJ et al (2007) The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14:869–871. https://doi.org/10.1038/nsmb1283
Zhang P, Zhu C, Geng Y et al (2021) Rice and Arabidopsis homologs of yeast chromosome transmission fidelity protein 4 commonly interact with Polycomb complexes but exert divergent regulatory functions. Plant Cell 33:1417–1429. https://doi.org/10.1093/plcell/koab047
Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG (2012) Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J Biol Chem 287:7859–7870. https://doi.org/10.1074/jbc.M111.336271
Acknowledgements
We sincerely thank Dr. Yuehui He (from School of Advanced Agricultural Sciences, Peking University) for kindly providing the prmt6, nf-yc3;4;9 mutants, and the NF-Y plasmids for Y2H assays, and also thank him for critically reading this manuscript.
Funding
This work was supported by the Natural National Science Foundation of China (32101786), the National Transgenic Major Program (2019ZX08010-002) and the Fundamental Research Funds for Central Non-profit Scientific Institution (1610392017001), and the Baichuan Project at the College of Life Science and Technology, Huazhong Agricultural University.
Author information
Authors and Affiliations
Contributions
LY supervised the study. PZ and XG conceived and designed the experiments. PZ performed the most experiments. XL, YW, WG, and YG partially performed the experiments. AR and SC contributed to the writing. PZ, XG, and LY wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest needs to be declared.
Additional information
The original online version of this article was revised: Figs 2 and 3 were wrongly numbered; Fig. 2 should have been Fig. 3 and vice versa.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, P., Li, X., Wang, Y. et al. PRMT6 physically associates with nuclear factor Y to regulate photoperiodic flowering in Arabidopsis. aBIOTECH 2, 403–414 (2021). https://doi.org/10.1007/s42994-021-00065-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-021-00065-y