Abstract
The RNAi technology takes advantage of the intrinsic RNA interference (RNAi) mechanism that exists in nearly all eukaryotes in which target mRNAs are degraded or functionally suppressed. Significant progress has been made in recent years where RNAi technology is applied to several crops and economic plants for protection against diseases like fungi, pests, and nematode. RNAi technology is also applied in controlling pathogen damages in wheat, one of the most important crops in the world. In this review, we first give a brief introduction of the RNAi technology and the underneath mechanism. We then review the recent progress of its utilization in crops, particular wheat. Finally, we discuss the existing challenges and prospect future development of this technology in crop protection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) contributes more than 20% of the total dietary calories and proteins for humans worldwide (Shiferaw et al. 2013). It plays a pivotal role in securing the global food demand. The increase of wheat yield, however, has slowed down in recent years partly due to newly emerging varieties of various diseases—pathogens, pests and nematodes (Rosegrant and Cline 2003). On the other hand, the overuse of pesticides for disease control has posed substantial risks to food safety, the environment, and all living organisms (Ali 2014). The transgenic crops expressing insecticidal proteins from Bacillus thuringiensis (Bt) effectively reduced the insecticide usage and increased crop yields. However, the limited scope of Bt crops and the appearance of Bt-resistant pests in fields call for new technologies for pest control (Carriere et al. 2015; Jin et al. 2015; Tabashnik et al. 2013). The phenomenon of RNA interference (RNAi) is widely found in eukaryotes (plants, fungi, insects, animals, and nematodes etc.) and has been developed as a promising technology for crop health protection (Zhang et al. 2017). RNAi is a natural process that involves the regulation of gene expression by several manners: effective post-transcriptional gene silencing (PTGS), translational inhibition, RNA destabilization, and/or transcriptional gene silencing (TGS) by directing DNA methylation (Fire et al. 1998; Coleman et al. 2015; Ghildiyal et al. 2008; Huvenne and Smagghe 2010; Jones-Rhoades et al. 2006; Liu et al. 2020; Mao et al. 2007; Sherman et al. 2015). Here, we review recent progress in the development of RNAi-based plant protection technologies, particularly on its application in wheat. We discuss its potential for the control of fungal pathogens, pests and nematodes, as well as current challenges facing RNAi strategy. We also prospect the future improvement in delivery methods for effective applications of this technology in crop protection.
The mechanism of RNAi technology
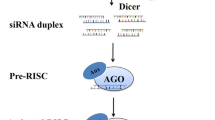
RNAi is a self-protection mechanism in eukaryotic cells and is triggered by double-stranded RNA (dsRNA) when present in a cell. dsRNA is processed by the ribonuclease III enzyme Dicer or Dicer-like enzymes to produce small interfering RNAs (siRNAs) of 20–30 nucleotide (nt) long. These small RNA (sRNA) are bound to Argonaute family proteins (AGOs), the catalytic components of the RNAi system. The AGO/siRNA complexes are then recruited to the RNA-induced silencing complex (RISC) (Lee et al. 2010), which mediates mRNA degradation, mRNA translation, or chromatin modification (Borges and Martienssen 2015) (Fig. 1). In most eukaryotes, including pathogens and pests, RNA-dependent RNA polymerases (RdRPs) have been identified for secondary dsRNA synthesis and are essential for the systemic effect of RNAi. Two works have specified functions for the RdRP activity in RNAi in Caenorhabditis elegans (Sijen et al. 2001) and fungi (Dang et al. 2011); however, a similar RdRP-based amplification system is yet to be discovered in insects (Zotti et al. 2018). Given the presence of RNAi pathways in pathogens, pests, and nematodes, it is not surprising to take advantage of its working mechanism in crop protection.
The RNAi technology and its application in crop diseases control. (Left) Crop disease control by RNAi. dsRNA delivery strategies for wheat protection mainly via HIGS, foliar sprays and recombinant microbes. Each of these strategies contains advantages, relying on the specific condition involved. Additional methods are also used such as nanoparticles, baits, trunk injection, and root soaking. (Right) The RNAi mechanism. Double-stranded RNA (dsRNA) molecule binds to a Dicer/Dcl protein, which cleaves it into small interfering RNAs (siRNAs); these siRNAs bind to an Argonaute (AGO) protein, to form an RNA-Induced Silencing Complex (RISC), then RISC separates the siRNAs into two strands. The siRNA/RISC complex binds the complementary sequence of the target mRNA resulting in post-transcriptional gene silencing (PTGS) via degradation of the mRNA target (indicated by a scissor) or inhibition of its translation (indicated by red vertical bar), or resulting in transcriptional gene silencing (TGS) in the nucleus via chromatin modifications. In fungi and nematodes, the silencing signal can be perpetuated by the action of the RNA-dependent RNA polymerase (RdRP) for siRNA secondary amplification, but for pests, RdRP is not yet found
Delivery of interfering RNAs
Interspecific transportation of sRNAs takes place naturally. siRNAs can be shuttled between plants and pathogens by secreted vesicles (Cai et al. 2018; Weiberg et al. 2013). In cotton, the production of microRNAs (miRNAs) miR166 and miR159 was increased upon Verticillium dahliae (a vascular fungal pathogen responsible for devastating wilt diseases in many crops) infection and transported to infection sites to silence virulence genes reducing its damage (Zhang et al. 2016). Despite these in vivo mechanisms, the RNAi technology is impeded by in vitro dsRNA delivering efficiency. Numerous efforts on artificial delivery methods have been attempted. The selection of the suitable delivery approaches (e.g. host-induced gene silencing, foliar sprays, recombinant microbes) is in fact determining the success of the technology (Fig. 1). A few methods have been tested.
The first approach is the application of synthetic dsRNA or sRNA derived from pathogen or pest genes as pesticides on crop leaves. Foliar application with sprayable RNAi-based products, such as sRNAs, is suitable for controlling pests and pathogens on stems, foliage, or fruits. The products are evaluated similarly to topical pesticides where the RNA solution is sprayed on leaves, or fed to the target pests, and impacts on insects are then observed (Andrade and Hunter 2017). One of the first case exploring the applications of sprayable RNA molecules to control pests was the use of siRNA against the diamondback moth (Plutella xylostella). Brassica leaves that were sprayed with chemically synthesized siRNAs targeting the acetylcholine esterase gene AchE2 caused high mortality for P. xylostella. (Gong et al. 2013). In another case, foliar application of dsRNA targeting the cytochrome P450 (CYP3) gene of Fusarium graminearum resulted in successful inhibition of fungal growth in directly sprayed leaves as well as the distal non-sprayed leaves in barley plants (Koch et al. 2016). This strategy or so-called spray-induced gene silencing (SIGS) opens an avenue of development of biopesticide which is environmentally friendly. Moreover, since RNAi is highly dependent on the sequence specificity, it has little effects on the non-target microorganisms or non-target pests.
The second method is to use recombinant microbes such as virus and bacteria engineered to produced dsRNA in host crops (Cagliari et al. 2018; Dubrovina and Kiselev 2019; Goulin et al. 2019). Virus-induced gene silencing (VIGS) is a naturally occurring (Baulcombe 2015; Waterhouse et al. 2001). Unlike stable RNAi and mutants, the transiently expressed dsRNA by VIGS does not modify plant genetic composition. For instance, three midgut-expressed CYP genes of the Lepidoptera insect, Manduca sexta were targeted through viral vectors to produce dsRNA in the host plant. The viral vector was engineered using Tobacco Rattle Virus (TRV) to deliver dsRNA into Nicotiana attenuata (Kumar et al. 2012). DsRNA could also be produced in the bacteria (HT115). When the cotton bollworm (Helicoverpa armigera) larvae fed with the artificial diet coated with dsRNA expressing HT115, high mortality was observed after five days. Data showed that inhibition of target gene expression led to significant reductions in body weight, body length, and pupation rate (Ai et al. 2018).
The third approach is host-induced gene silencing (HIGS) which employs transgenic plants to produce dsRNA derived from pathogen or pest genes. RNAi occurs in pests when they ingest sufficient dsRNA or sRNA. Tests have been made for a few pests where persistent effects were obtained for several common species (Baum et al. 2007; Mao and Zeng 2014; Sun et al. 2019; Zhu et al. 2012). The phloem-feeding hemipterans such as aphids with specialized mouthparts (stylets) that penetrate through plant tissues to ingest cell saps. In this case, dsRNA sequences of shp gene effectively reduced the growth, the reproduction, and the survival rate of tested aphids. Remarkably, other developmental aberrations were also observed such as winged adults and delayed maturation (Abdellatef et al. 2015). This method is a complementary tool to Bt-based insect-resistant plants which is not effective for several hemipterans with specialized stylets. Cotton plants constitutively expressing dsRNA from genes encoding the P450 protein CYP6AE14 and NDPH dehydrogenase protein 2 of cotton bollworm (Helicoverpa armigera) significantly improved resistance to this pest, and the dsNDPH cotton is almost equivalent to Bt cottons in resistance efficiency (Mao et al. 2011; Wu et al. 2016). Similarly, dsRNA homologous to V-type ATPase gene of corn root worm (Diabrotica virgifera) in transgenic corn plants rendered significant improvement of insect resistance (Baum et al. 2007).
For woody plants, such as fruit trees, dsRNA can be delivered via insecticidal baits, nanoparticle trunk injection and root soaking. The information of these methods can be found elsewhere for detail (Liu et al. 2020; Zhu and Palli 2019).
The application of RNAi for wheat protection
Management of bacterial and fungal pathogens
In wheat, a few serious wheat diseases, such as Fusarium head blight (FHB) caused by necrotrophic fungi of the genus Fusarium and leaf rust caused by biotrophic fungi of the genus Puccinia (Table 1), have been targeted using RNAi technology. Transgenic wheat plants were engineered to confer three hairpin RNA fragments derived from the Fusarium graminearum chitin synthase gene (Chs3b), which is responsible for the biosynthesis of chitin. These transgenic plants showed strong resistance to FHB and Fusarium seedling blight (FSB) (Cheng et al. 2015). On the other hand, expressing dsRNA complementary to mRNAs of Puccinia triticina MAP-kinase (PtMAPK1, 520 bp) or a cyclophilin (PtCYC1, 501 bp) showed efficient silencing of the corresponding genes in the fungus and significant reduction of the fungal pathogenicity and growth in transgenic wheat. P. triticina is an aggressive fungal pathogen that causes severe leaf rust disease in wheat. P. triticina proliferation was significantly reduced together with decreasing fungal target gene transcript abundance and reduced biomass accumulation in RNAi-based resistant plants (Panwar et al. 2018).
Powdery mildew caused by Blumeria graminis f. sp. hordei in barley and B. graminis f. sp. tritici in wheat is a serious disease as well. Transgenic barley expressing dsRNA targeting the avirulence gene Avra10, which corresponds to the resistance gene Mla10, showed reduced fungal gene transcripts and severely affected fungal development (Nowara et al. 2010). Silencing of 1,3-β-glucanosyltransferase genes (BgGTF1 and BgGTF2) via VIGS that was built on the barley stripe mosaic virus (BSMV) significantly slowed down the growth of the powdery mildew fungus (Qi et al. 2019b). Mildew resistance locus o (Mlo) encodes a transmembrane protein (Panstruga et al. 2005) that acts as a negative regulator to suppress plant immunity in uninfected tissues. It is also involved in protection against cell death as well as in responses to biotic and abiotic stresses (Piffanelli et al. 2002). Down-regulation of the TaMlo gene via VIGS resulted in the broad-spectrum powdery mildew resistance in wheat (Várallyay et al. 2012). Recently, gene-editing technologies were used to achieve similar effects. For instance, simultaneous knockout of the three TaMlo homoeologues by TALEN (transcription activator‐like effector nuclease) produced transgenic wheat plants that were highly resistant to powdery mildew infection, another work produced transgenic wheat plants that carry mutations in the TaMLO-A1 allele using the CRISPR-Cas9 technology (Wang et al. 2014). On the other hand, non‐transgenic TILLING (Targeting Induced Lesions IN Genomes) plants with partial loss‐of‐function alleles of TaMlo confer durable broad‐spectrum powdery mildew resistance (Acevedo-Garcia et al. 2017).
Wheat streak mosaic virus (WSMV) is another persistent threat to wheat production. Transgenic wheat plants constitutively expressing a polycistronic cassette of five miR395 arms, known as FanGuard (FGmiR395), were exploited to target five distinct regions of the virus genome. The consequent transgenic plants showed nearly complete immunity to WSMV (Fahim et al. 2012). In the other case, a segment of 272 bp sequence derived from the coat protein of Triticum mosaic virus (TriMV) was cloned into the hairpin expression vector and constitutively expressed in wheat. The engineered wheat plants showed stable resistance to TriMV (Fahim et al. 2010).
Management of wheat pests
Several major pests, such as grain aphid (Sitobion avenae), bird cherry-oat aphid (Rhopalosiphum padi), and wheat aphid (Schizaphis graminum), can cause severe yield loss (Table 1) (Peairs 2008; Smith and Chuang 2014; Yu et al. 2014). Transgenic wheat plants expressing a 198 bp fragment of dsRNA complementary to the zinc finger protein (SaZFP) of grain aphid can effectively increase its mortality and reduce its daily fecundity (Sun et al. 2019). In barley, dsRNA targeting the grain aphid gene encoding salivary sheath protein (SHP), a pivotal component of the stylet penetration process, effectively reduces the reproduction and survival rates of the aphid and the effect can be transmitted for seven generations (Abdellatef et al. 2015). Effects of additional target genes were also confirmed by feeding or direct injection into grain aphid, such as those encoding catalase, acetylcholinesterase1, cytochrome c oxidase subunit VIIc precursor, and zinc finger protein, and abnormally high mortality and developmental stunting were observed (Wang et al. 2015; Zhang et al. 2013).
Management of nematodes in wheat
Wheat parasites cause enormous yield losses and threaten the quality of grains, including Heterodera avenae, H. filipjevi and H. latipons (Table 1) (Toumi and Waeyenberge 2013). Targeting of the Ha18764 effector protein family genes of H. avenae by the VIGS-based RNAi approach significantly attenuated the parasitism and reproduction status of H. avenae in wheat (Yang et al. 2019). Down-regulation by RNAi of pat-10 and unc-87 genes on Thorne's meadow nematode (Pratylenchus thornei), which infects wheat roots, significantly reduced the reproduction of the worms (Tan et al. 2013). Moreover, RNAi in wheat can be stimulated by poly-component biostimulants derived from metabolites of various soil streptomycetes which up-regulate siRNAs and miRNAs in wheat plants. These small RNAs are complementary to cereal cyst nematode mRNA and hence suppress their reproduction providing resistance to wheat plants (Blyuss et al. 2019).
Challenges for using RNAi technology
While the outlook of using RNAi for plant protection appears to be promising, several issues need to be resolved before efficient practical applications.
The stability of dsRNA
One of the primary concerns for the use of RNA as a biopesticide is their stability, especially for the sprayable dsRNA and siRNA applications. Microorganisms in the environment can degrade dsRNA prior to their uptake by pathogens or pests. Rapid degradation of dsRNA may occur by nucleases in the saliva, gut lumen, and/or haemolymph of pests as well (Allen and Walker III 2012; Chung et al. 2018; CoGuan et al. 2018; Katoch and Thakur 2012; Kennedy et al. 2004; Luo et al. 2013). The high or low pH found in the gut lumens of some pests can also reduce dsRNA stability either directly or indirectly by affecting the activity of gut nucleases (Cooper et al. 2019).
Other environmental factors may exert different effects on the stability of dsRNA and sRNA. Several works show that dsRNA is degraded to undetectable levels within 48 h after their application on three types of soil (silt loam, loamy sand, and clay loam) and within 7 days after their addition to aquatic systems containing natural water and various types of sediment (Albright et al. 2017; Fischer et al. 2017). Despite this, actin-dsRNA derived Colorado potato beetle (CPB) remained active for at least four weeks after application to potato leaves. It suppressed CPB larval weight gain, delayed its development, and increased its mortality (San Miguel and Scott 2016). Therefore, dissecting the process of dsRNA degradation is helpful in evaluating the potential effect of dsRNA in various environments and target organisms.
Cost-effective methods for dsRNA production
For RNAi application to be practical for field use, the major hurdle is to produce sufficient amount of dsRNA. The traditional dsRNA production method in the laboratory is expensive and produces only a limited amount of dsRNA and thus is not practical for large-scale application needs (Ahn et al. 2019). Producing dsRNA in bacterial cells with RNaseIII deficiency seems to be an alternative. However, only a handful works have demonstrated microbial-based dsRNA production. One approach uses L1440-HT115 (DE3) system that has been successfully applied in the RNAi of Mythimna separate (Das et al. 2015; Parsons et al. 2018; Zhang et al. 2010). With more research underway, the production efficiency of this system should be augmented to meet market demands.
Off-target effects
RNAi is a sequence homology-dependent mechanism. Several studies show that siRNA is not always specific and can have off-target effects and thus is problematic in disease management (Mamta and Rajam 2017). Some target genes are highly conserved between species which increases the likelihood of off-targets among them. The sequences of vATPaseA and vATPaseEfrom L. decemlineata, for instance, shared 83% and 79% nucleotide-sequence identities to their counterparts in Western Corn Rootworm (WCR), respectively. dsRNAs from WCR vATPaseA and vATPaseE could reduce the fitness of Colorado potato beetle (CPB; Leptinotarsa decemlineata) in a bioassay (Baum et al. 2007). Computational design program is needed for specific and systemic evaluation of non-target and off-target effects which should be further verified by additional bioassays. In addition, feeding studies revealed that dsRNAs of at least 60 nucleotide (nt) in length are necessary for an efficient RNAi response in D. virgifera (Bolognesi et al. 2012) and Tribolium castaneum (Wang et al. 2019). A minimum of 21 nt was required for the size of siRNA for efficient protection against WCR and active orthologs (Bachman et al. 2013).
RNAi resistance
Pests and pathogens can develop resistance to RNAi-based products through various mechanisms as they do for conventional biopesticides. Compared to conventional commercialized transgenic crops expressing Bt toxins for pest management (James, 2010). RNAi-based strategy induces down-regulation of the target gene by in-complete resistance in most of cases. This may reduce the selection pressure on the pathogen that may contribute to durable resistance. But genetic variation in pathogenic organisms may also cause single nucleotide polymorphisms (SNPs) in the target gene. The efficiency of RNAi would be cut down owing to the reduction of complementarity between the target gene and the dsRNA. Synonymous SNPs lead to nearly no fitness cost on the pathogens and pests, but the difference between dsRNA and the original gene sequences reduces their complementarity, causing reduced RNAi effect or RNAi resistance (Scott et al. 2013; Yu et al. 2016). Thereby, the potential of RNAi resistance should be taken into consideration in application.
Conclusions and future prospects
In the past few years, we have seen diverse applications of RNAi in crop protection methodologies against pests, pathogens, and nematodes. RNAi technology has emerged as a promising new strategy for wheat protection either. The wide use of HIGS on a commercial scale appears possible soon. The major obstacles for the HIGS strategy will be resolved, by optimal target and fragment selection methods, highly efficient transformation constructs, and stable transgenic systems. To this end, it is worthy to mention that the V-type ATPase-based RNAi technology has passed the GM safety evaluation in eight countries and regions including the United States, Brazil, and Japan. It has also been licensed for planting by the US Environmental Protection Agency (Zotti et al. 2018), painting an elusive picture for the commercialization of the RNAi technology. Technical barriers are being overcome to allow a wide range of applications from laboratory to the field. The technology of encapsulated dsRNA on leaves with SIGS has significantly promoted dsRNA stability in the environment as well as during its uptake by pests enhancing plant protection. Cost-effective approaches for massive production of dsRNA (e.g. bacterial, plant, and synthetic production) are being optimized and will contribute to lowering costs of the technology. There is no doubt that a new era of disease control based on RNAi technology for crop protection is right at the corner.
References
Abdellatef E, Will T, Koch A, Imani J, Vilcinskas A, Kogel KH (2015) Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol J 13(6):849–857. https://doi.org/10.1111/pbi.12322
Acevedo-Garcia J, Spencer D, Thieron H, Reinstädler A, Hammond-Kosack K, Phillips AL, Panstruga R (2017) mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol J 15(3):367–378. https://doi.org/10.1111/pbi.12631
Ahn SJ, Donahue K, Koh Y, Martin RR, Choi MY (2019) Microbial-based double-stranded RNA production to develop cost-effective RNA interference application for insect pest management. Int J Insect Sci 11:1179543319840323. https://doi.org/10.1177/1179543319840323
Ai X, Wei Y, Huang L, Zhao J, Wang Y, Liu X (2018) Developmental control of Helicoverpa armigera by ingestion of bacteria expressing dsRNA targeting an arginine kinase gene. Biocontrol Sci Tech 28:253–267. https://doi.org/10.1080/09583157.2018.1441368
Albright VC 3rd, Wong CR, Hellmich RL, Coats JR (2017) Dissipation of double-stranded RNA in aquatic microcosms. Environ Toxicol Chem 36(5):1249–1253. https://doi.org/10.1002/etc.3648
Ali P (2014) Pesticide overuse: stop killing the beneficial agents. J Environ Anal Toxicol 4. https://doi.org/10.4172/2161-0525.1000223
Allen ML, Walker WB III (2012) Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J Insect Physiol 58(3):391–396. https://doi.org/10.1016/j.jinsPhys.2011.12.014
Andrade EC, Hunter WB (2017) RNAi feeding bioassay: development of a non-transgenic approach to control Asian citrus psyllid and other hemipterans. Entomol Exp Appl 162:389–396. https://doi.org/10.1111/eea.12544
Bachman PM et al (2013) Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res 22(6):1207–1222. https://doi.org/10.1007/s11248-013-9716-5
Baulcombe DC (2015) VIGS, HIGS and FIGS: small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr Opin Plant Biol 26:141–146. https://doi.org/10.1016/j.pbi.2015.06.007
Baum JA et al (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25(11):1322–1326. https://doi.org/10.1038/nbt1359
Blyuss KB, Fatehi F, Tsygankova VA, Biliavska LO, Iutynska GO, Yemets AI, Blume YB (2019) RNAi-based biocontrol of wheat nematodes using natural poly-component biostimulants. Front Plant Sci 10:483. https://doi.org/10.3389/fpls.2019.00483
Bolognesi R et al (2012) Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 7:e47534. https://doi.org/10.1371/journal.pone.0047534
Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16:727–741. https://doi.org/10.1038/nrm4085
Cagliari R, Kremer FS, da Silva PL (2018) Bauhinia lectins: biochemical properties and biotechnological applications. Int J Biol Macromol 119:811–820. https://doi.org/10.1016/j.ijbiomac.2018.07.156
Cai Q et al (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360(6393):1126–1129. https://doi.org/10.1126/science.aar4142
Carriere Y, Crickmore N, Tabashnik BE (2015) Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol 33:161–168. https://doi.org/10.1038/nbt.3099
Chen W et al (2016) Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J Exp Bot 67:4979–4991. https://doi.org/10.1093/jxb/erw263
Cheng W et al (2015) Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol J 13:1335–1345. https://doi.org/10.1111/pbi.12352
Chung SH, Jing X, Luo Y, Douglas AE (2018) Targeting symbiosis-related insect genes by RNAi in the pea aphid-Buchnera symbiosis. Insect Biochem Mol Biol 95:55–63. https://doi.org/10.1016/j.ibmb.2018.02.004
Coleman AD, Wouters RH, Mugford ST, Hogenhout SA (2015) Persistence and transgenerational effect of plant-mediated RNAi in aphids. J Exp Bot 66(2):541–548. https://doi.org/10.1093/jxb/eru450
Cooper AM, Silver K, Zhang J, Park Y, Zhu KY (2019) Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag Sci 75:18–28. https://doi.org/10.1002/ps.5126
Dang Y, Yang Q, Xue Z, Liu Y (2011) RNA interference in fungi: pathways, functions, and applications. Eukaryot Cell 10:1148. https://doi.org/10.1128/EC.05109-11
Das S, Debnath N, Cui Y, Unrine J, Palli SR (2015) Chitosan, carbon quantum dot, and silica nanoparticle mediated dsRNA delivery for gene silencing in Aedes aegypti: a comparative analysis. ACS Appl Mater Interfaces 7:19530–19535. https://doi.org/10.1021/acsami.5b05232
Deng F, Zhao Z (2014) Influence of catalase gene silencing on the survivability of Sitobion avenae. Arch Insect Biochem Physiol 86:46–57. https://doi.org/10.1002/arch.21161
Dubrovina AS, Kiselev KV (2019) Exogenous RNAs for gene regulation and plant resistance. Int J Mol Sci. https://doi.org/10.3390/ijms20092282
Fahim M, Ayala-Navarrete L, Millar AA, Larkin PJ (2010) Hairpin RNA derived from viral NIa gene confers immunity to wheat streak mosaic virus infection in transgenic wheat plants. Plant Biotechnol J 8:821–834. https://doi.org/10.1111/j.1467-7652.2010.00513.x
Fahim M, Millar AA, Wood CC, Larkin PJ (2012) Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol J 10:150–163. https://doi.org/10.1111/j.1467-7652.2011.00647.x
Fan J, Zhang Y, Francis F, Cheng D, Sun J, Chen J (2015) Orco mediates olfactory behaviors and winged morph differentiation induced by alarm pheromone in the grain aphid, Sitobion avenae. Insect Biochem Mol Biol 64:16–24. https://doi.org/10.1016/j.ibmb.2015.07.006
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. https://doi.org/10.1038/35888
Fischer JR et al (2017) Aquatic fate of a double-stranded RNA in a sediment–-water system following an over-water application. Environ Toxicol Chem 36:727–734. https://doi.org/10.1002/etc.3585
Ghildiyal M et al (2008) Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320:1077–1081. https://doi.org/10.1002/etc.3585
Gong L, Chen Y, Hu Z, Hu M (2013) Testing insecticidal activity of novel chemically synthesized siRNA against Plutella xylostella under laboratory and field conditions. PLoS ONE 8:e62990–e62990. https://doi.org/10.1371/journal.pone.0062990
Goulin EH, Galdeano DM, Granato LM, Matsumura EE, Dalio RJD, Machado MA (2019) RNA interference and CRISPR: promising approaches to better understand and control citrus pathogens. Microbiol Res 226:1–9. https://doi.org/10.1016/j.micres.2019.03.006
Guan RB, Li HC, Fan YJ, Hu SR, Christiaens O, Smagghe G, Miao XX (2018) A nuclease specific to lepidopteran insects suppresses RNAi. J Biol Chem 293:6011–6021. https://doi.org/10.1074/jbc.RA117.001553
Huvenne H, Smagghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56:227–235. https://doi.org/10.1016/j.jinsphys.2009.10.004
James C (2010) A global overview of biotech (GM) crops: adoption, impact and future prospects. GM Crops 1:8–12. https://doi.org/10.4161/gmcr.1.1.9756
Jin L, Zhang H, Lu Y, Yang Y, Wu K, Tabashnik BE, Wu Y (2015) Large-scale test of the natural refuge strategy for delaying insect resistance to transgenic Bt crops. Nat Biotechnol 33:169–174. https://doi.org/10.1038/nbt.3100
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53. https://doi.org/10.1146/annurev.arplant.57.032905.105218
Katoch R, Thakur N (2012) Insect gut nucleases: a challenge for RNA interference mediated insect control strategies. Int J Biochem Biotechnol 1:198–203
Kennedy S, Wang D, Ruvkun G (2004) A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427:645–649. https://doi.org/10.1038/nature02302
Koch A et al (2016) An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog 12:e1005901–e1005901. https://doi.org/10.1371/journal.ppat.1005901
Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel K-H (2013) Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc Natl Acad Sci USA 110(48):19324–19329. https://doi.org/10.1073/pnas.1306373110
Kumar P, Pandit SS, Baldwin IT (2012) Tobacco rattle virus vector: a rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS ONE 7:e31347. https://doi.org/10.1371/journal.pone.0031347
Lee HC et al (2010) Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell 38:803–814. https://doi.org/10.1016/j.molcel.2010.04.005
Lilley CJ, Bakhetia M, Charlton WL, Urwin PE (2007) Recent progress in the development of RNA interference for plant parasitic nematodes. Mol Plant Pathol 8:701–711. https://doi.org/10.1111/j.1364-3703.2007.00422.x
Liu S, Jaouannet M, Dempsey DMA, Imani J, Coustau C, Kogel K-H (2020) RNA-based technologies for insect control in plant production. Biotechnol Adv 39:107463. https://doi.org/10.1016/j.biotechadv.2019.107463
Luo Y, Wang X, Wang X, Yu D, Chen B, Kang L (2013) Differential responses of migratory locusts to systemic RNA interference via double-stranded RNA injection and feeding. Insect Mol Biol 22:574–583. https://doi.org/10.1111/imb.12046
Mamta B, Rajam MV (2017) RNAi technology: a new platform for crop pest control. Physiol Mol Biol Plants 23:487–501. https://doi.org/10.1007/s12298-017-0443-x
Mao J, Zeng F (2014) Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res 23:145–152. https://doi.org/10.1007/s11248-013-9739-y
Mao YB et al (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25:1307–1313. https://doi.org/10.1038/nbt1352
Mao YB, Tao XY, Xue XY, Wang LJ, Chen XY (2011) Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res 20:665–673. https://doi.org/10.1007/s11248-010-9450-1
Nowara D et al (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22:3130–3141. https://doi.org/10.1105/tpc.110.077040
Panstruga R (2005) Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol Biol 59:485–500. https://doi.org/10.1007/s11103-005-0353-0
Panwar V, Jordan M, McCallum B, Bakkeren G (2018) Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol J 16:1013–1023. https://doi.org/10.1111/pbi.12845
Panwar V, McCallum B, Bakkeren G (2013) Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol Biol 81:595–608. https://doi.org/10.1007/s11103-013-0022-7
Parsons KH, Mondal MH, McCormick CL, Flynt AS (2018) Guanidinium-functionalized interpolyelectrolyte complexes enabling RNAi in resistant insect pests. Biomacromol 19:1111–1117. https://doi.org/10.1021/acs.biomac.7b01717
Peairs FB (2008) Wheat pests and their management. In: Capinera JL (ed) Encyclopedia of entomology. Springer, Netherlands, pp 4220–4238. https://doi.org/10.1007/978-1-4020-6359-6_2660
Piffanelli P et al (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129:1076–1085. https://doi.org/10.1104/pp.010954
Pliego C et al (2013) Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol Plant Microbe Interact 26:633–642. https://doi.org/10.1094/mpmi-01-13-0005-r
Qi T, Guo J, Liu P et al (2019a) Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-Promoting transcription factor TaLOL2 to defeat ROS-Induced defense in wheat. Mol Plant 12(12):1624–1638. https://doi.org/10.1016/j.molp.2019.09.010
Qi T, Guo J, Peng H, Liu P, Kang Z, Guo J (2019b) Host-induced gene silencing: a powerful strategy to control diseases of wheat and barley. Int J Mol Sci 20:206. https://doi.org/10.3390/ijms20010206
Qi T, Zhu X, Tan C, Liu P, Guo J, Kang Z, Guo J (2018) Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol J 16:797–807. https://doi.org/10.1111/pbi.12829
Rosegrant MW, Cline SA (2003) Global food security: challenges and policies. Science 302(5652):1917–1919. https://doi.org/10.1126/science.1092958
San Miguel K, Scott JG (2016) The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag Sci 72:801–809. https://doi.org/10.1002/ps.4056
Schaefer LK et al (2020) Cross-kingdom RNAi of pathogen effectors leads to quantitative adult plant resistance in wheat. Front Plant Sci. https://doi.org/10.3389/fpls.2020.00253
Scott JG et al (2013) Towards the elements of successful insect RNAi. J Insect Physiol 59:1212–1221. https://doi.org/10.1016/j.jinsphys.2013.08.014
Sherman JH, Munyikwa T, Chan SY, Petrick JS, Witwer KW, Choudhuri S (2015) RNAi technologies in agricultural biotechnology: the toxicology forum 40th annual summer meeting. Regul Toxicol Pharmacol 73:671–680. https://doi.org/10.1016/j.yrtph.2015.09.001
Shiferaw B, Smale M, Braun H-J, Duveiller E, Reynolds M, Muricho G (2013) Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Sec 5:291–317. https://doi.org/10.1007/s12571-013-0263-y
Sijen T et al (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465–476
Smith CM, Chuang WP (2014) Plant resistance to aphid feeding: behavioral, physiological, genetic and molecular cues regulate aphid host selection and feeding. Pest Manag Sci 70:528–540. https://doi.org/10.1002/ps.3689
Song XS et al (2018) A myosin5 dsRNA that reduces the fungicide resistance and pathogenicity of Fusarium asiaticum. Pestic Biochem Physiol 150:1–9. https://doi.org/10.1016/j.pestbp.2018.07.004
Sun Y, Sparks C, Jones H, Riley M, Francis F, Du W, Xia L (2019) Silencing an essential gene involved in infestation and digestion in grain aphid through plant-mediated RNA interference generates aphid-resistant wheat plants. Plant Biotechnol J 17:852–854. https://doi.org/10.1111/pbi.13067
Tabashnik BE, Brévault T, Carrière Y (2013) Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol 31:510–521. https://doi.org/10.1038/nbt.2597
Tan J-ACH, Jones MGK, Fosu-Nyarko J (2013) Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp Parasitol 133:166–178. https://doi.org/10.1016/j.exppara.2012.11.011
Toumi F, Waeyenberge L (2013) Development of two species-specific primer sets to detect the cereal cyst nematodes Heterodera avenae and Heterodera filipjevi. Eur J Plant Pathol 136:613–624. https://doi.org/10.1007/s10658-013-0192-9
Várallyay É, Giczey G, Burgyán J (2012) Virus-induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch Virol 157:1345–1350. https://doi.org/10.1007/s00705-012-1286-y
Wang D, Liu Q, Li X, Sun Y, Wang H, Xia L (2015) Double-stranded RNA in the biological control of grain aphid (Sitobion avenae F.). Funct Integr Genomics 15:211–223. https://doi.org/10.1007/s10142-014-0424-x
Wang K, Peng Y, Fu W, Shen Z, Han Z (2019) Key factors determining variations in RNA interference efficacy mediated by different double-stranded RNA lengths in Tribolium castaneum. Insect Mol Biol 28:235–245. https://doi.org/10.1111/imb.12546
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J-L (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951. https://doi.org/10.1038/nbt.2969
Waterhouse PM, Wang M-B, Lough T (2001) Gene silencing as an adaptive defence against viruses. Nature 411:834–842. https://doi.org/10.1038/35081168
Weiberg A et al (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342:118–123. https://doi.org/10.1126/science.1239705
Wu XM, Yang CQ, Mao YB, Wang LJ, Shangguan XX, Chen XY (2016) Targeting insect mitochondrial complex I for plant protection. Plant Biotechnol J 14:1925–1935. https://doi.org/10.1111/pbi.12553
Xiao D, Lu YH, Shang QL, Song DL, Gao XW (2015) Gene silencing of two acetylcholinesterases reveals their cholinergic and non-cholinergic functions in Rhopalosiphum padi and Sitobion avenae. Pest Manag Sci 71:523–530. https://doi.org/10.1002/ps.3800
Xu L, Hou Q, Zhao Y, Lu L, Li B, Ni Z, Liang R (2017) Silencing of a lipase maturation factor 2-like gene by wheat-mediated RNAi reduces the survivability and reproductive capacity of the grain aphid. Sitobion avenae Arch Insect Biochem Physiol 95:e21392. https://doi.org/10.1002/arch.21392
Yan T, Chen H, Sun Y, Yu X, Xia L (2016) RNA interference of the ecdysone receptor genes EcR and USP in grain Aphid (Sitobion avenae F.) affects its survival and fecundity upon feeding on wheat plants. Int J Mol Sci 17(12):2098. https://doi.org/10.3390/ijms17122098
Yang S, Dai Y, Chen Y, Yang J, Yang D, Liu Q, Jian H (2019) A novel G16B09-like effector from Heterodera avenae suppresses plant defenses and promotes parasitism. Front Plant Sci 10:66. https://doi.org/10.3389/fpls.2019.00066
Yin C, Jurgenson JE, Hulbert SH (2010) Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. Tritici Mol Plant Microbe Interact 24:554–561. https://doi.org/10.1094/MPMI-10-10-0229
Yu X, Wang G, Huang S, Ma Y, Xia L (2014) Engineering plants for aphid resistance: current status and future perspectives. Theor Appl Genet 127(10):2065–2083. https://doi.org/10.1007/s00122-014-2371-2
Yu X-D et al (2016) RNAi-mediated plant protection against aphids. Pest Manag Sci 72:1090–1098. https://doi.org/10.1002/ps.4258
Zhang J, Khan SA, Heckel DG, Bock R (2017) Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol 35:871–882. https://doi.org/10.1016/j.tibtech.2017.04.009
Zhang M et al (2013) Identifying potential RNAi targets in grain aphid (Sitobion avenae F.) based on transcriptome profiling of its alimentary canal after feeding on wheat plants. BMC Genomics 14:560. https://doi.org/10.1186/1471-2164-14-560
Zhang T, Jin Y, Zhao JH, Gao F, Zhou BJ, Fang YY, Guo HS (2016) Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae. Mol Plant 9:939–942. https://doi.org/10.1016/j.molp.2016.02.008
Zhang X, Zhang J, Zhu KY (2010) Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol Biol 19:683–693. https://doi.org/10.1111/j.1365-2583.2010.01029.x
Zhang Y, Fan J, Francis F, Chen J (2018) Molecular characterization and gene silencing of Laccase 1 in the grain aphid, Sitobion avenae. Arch Insect Biochem Physiol 97:e21446. https://doi.org/10.1002/arch.21446
Zhao Y et al (2018) Plant-mediated RNAi of grain aphid CHS1 gene confers common wheat resistance against aphids. Pest Manag Sci 74:2754–2760. https://doi.org/10.1002/ps.5062
Zhu J-Q et al (2012) Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS ONE 7:e38572. https://doi.org/10.1371/journal.pone.0038572
Zhu KY, Palli SR (2019) Mechanisms, applications, and challenges of insect RNA interference. Annu Rev Entomol. https://doi.org/10.1146/annurev-ento-011019-025224
Zotti M, Dos Santos EA, Cagliari D, Christiaens O, Taning CNT, Smagghe G (2018) RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag Sci 74:1239–1250. https://doi.org/10.1002/ps.4813
Acknowledgements
We are grateful for the financial support by the National Natural Science Foundation of China (No. 31701429), National Key Program for Transgenic Research (No. 2016ZX08009-001), and the ‘Yellow Delta Scholarship’ from the municipal government of Dongying.
Author information
Authors and Affiliations
Contributions
LM and SSL conceived the topic. S.S.L drafted the first manuscript. SFG and ALL read the draft and gave substantial suggestions for revision, LM revised the manuscript with the help from YBM
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest.
Ethical approval
We declare that all materials and methods comply with required ethical standards.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, S., Geng, S., Li, A. et al. RNAi technology for plant protection and its application in wheat. aBIOTECH 2, 365–374 (2021). https://doi.org/10.1007/s42994-021-00036-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-021-00036-3