Abstract
Anthropogenic habitat fragmentation and roadkill mortality are considered important threats to European hedgehogs. Habitat fragmentation isolates hedgehog populations and, as a consequence, reduces their genetic diversity and leads the populations to vulnerable situations. The hedgehog populations in the Iberian Peninsula represent the southern limit of the species. We used microsatellite markers to estimate the genetic diversity and population structure of Erinaceus europaeus on the Iberian Peninsula. The obtained results indicated the presence of two differentiated groups, north-western and north-eastern, which coincided with the distribution of the two phylogeographic mitochondrial lineages described in the Peninsula. Moreover, in the north-eastern group, three genetically different clusters (Girona, Central Catalonia and Zoo) were identified. The highest genetic diversity (Hs = 0.696) was detected in the north-western region. Significant genetic differentiation (FST range = 0.072–0.224) was found among the clusters, indicating that these groups are well differentiated and present low gene flow. We concluded that the north-western group is genetically stable, whereas in the north-eastern region, despite some contact among groups, some populations are isolated and vulnerable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Western European hedgehog, Erinaceus europaeus L. (Erinaceidae: Erinaceinae), is naturally distributed in the Iberian Peninsula, western and central Europe, the UK, Ireland, Scandinavia and northern Russia (He et al. 2012). Western European hedgehogs are a nocturnal species inhabiting lowlands, deciduous woodlands and Mediterranean holm oak stands. They avoid dry regions but can be found in urban landscapes. Their distribution seems to be related to resource availability and the density of predators (Doncaster 1992; Micol et al. 1994; Young et al. 2006). The preference of hedgehogs for urban areas has been associated with the minor presence of badgers (Meles meles L.) and a major food availability in urban zones (Doncaster et al. 2001; Hof and Bright 2010; Pettett et al. 2017; De Poel et al. 2015). Morris (1988) concluded that hedgehogs usually do not move distances larger than 4 km, whereas Zingg (1994) found that some males can occasionally cover distances up to 7 km. Despite Reeve (1994) described juvenile dispersal, Rasmussen et al. (2019a) proved that in suburban areas hedgehogs have small home ranges mainly in their juvenile stage. Doncaster et al. (2001) indicated that hedgehogs do not have a clearly defined period of dispersion during their life history. These authors also stated that hedgehogs explore new territories and move from unfavourable to favourable locations, following variable routes with different lengths and directions, and reported dispersal movements up to 3.8 km. It seems that road verges could be used as dispersal corridors by hedgehogs, as has been described for other species of small mammals (Forman and Alexander 1998). Alternatively, roadways have also been reported to be significant barriers to hedgehog migration (Huijser and Bergers 2000; Orlowski and Nowak 2004).
Because gene flow among populations is expected to be correlated with the dispersion capabilities of their individuals (Allendorf 1983), knowledge of the possible genetic structure patterns can clarify the dispersion capabilities of hedgehogs. In this context, based on microsatellite genotyping, previous papers have assessed the connectivity of hedgehogs across highly fragmented habitats in the urban landscapes of Oxfordshire, UK (Becher and Griffiths 1998), Zurich, Switzerland (Braaker et al. 2017) and Berlin, Germany (Barthel et al. 2020). All these studies found high genetic diversity (HS = 0.6–0.7) and significant genetic differentiation among populations (FST = 0.06–0.19) explained by low rates of dispersal and landscape elements, such as rivers, highways or fences that hindered gene flow. In contrast, the recent study by Osaka et al. (2021) revealed low genetic diversity (HS = 0.39) and failed to detect population subdivision in urban hedgehogs around Helsinki. Interestingly, the study by Barthel et al. (2020) showed that the population structure disappeared when related individuals were excluded from the analysis. Microsatellite genotyping has also been used to study the biological invasion process of E. europaeus as an allochthonous species in New Zealand (Bolfíková et al. 2013) and Pianosa Island, Italy (Iannucci et al. 2019). In Denmark, using single nucleotide polymorphisms (SNPs) genotyping, Rasmussen et al. (2019b; 2020) found low genetic diversity values (HS = 0.13–0.32) and high population differentiation (FST = 0.03–0.32) across fragmented rural and urban landscapes, related with the isolated island structure of this country. Finally, other previous population genetic studies have focused on the contact zone of E. europaeus and E. roumanicus in Central Europe, where the pattern of population structure seems to be related to the postglacial colonization of the Iberian and Apennine phylogeographic lineages into this area (Bolfíková and Hulva 2012; Ploi et al. 2020; Zolotareva et al. 2021).
Other studies using molecular data have focused on the phylogeny and geographical distribution of E. europaeus evolutionary lineages, including none or a few specimens from the Iberian Peninsula (Santucci et al. 1998; Seddon et al. 2001; He et al. 2012; Bannikova et al. 2014). Recently, Sanz et al. (2021) studied the phylogeny of Iberian hedgehogs and found two western European mitochondrial lineages whose distribution was delimited by the Ebro River. However, no other studies exist regarding the population genetic or conservation status of hedgehog populations on the Iberian Peninsula, which represents the southern limit of the distribution of this species.
Declines in hedgehog densities have been reported in both rural and urban areas in Europe (Hof and Bright 2016; Williams et al. 2018; Taucher et al. 2020). The highest population decline has been documented in the rural areas of UK (Williams et al. 2018), where E. europaeus has been included as vulnerable in the Red List for Britains’ Mammals of 2020 (Mathews and Harrowers 2020). E. europaeus is listed in Appendix III of the Bern Convention and in the studied area (Spain) this species is protected by special conservation regulations of autonomic administrations (Catalonia, Valencia and Andalucia). However, E. europaeus is considered of least concern by the IUCN Red List of Threatened Species (IUCN 2021). The conservation of this species is also concerning in urban environments, which are considered to be critical areas to support global biodiversity (Aronson et al. 2017). The main important threats to hedgehog populations include fragmentation of their habitats due to human actions, presence of predators and road collisions (Huijser and Bergers 2000; Palomo et al. 2007; Hof and Bright 2009). The lack of connectivity among populations leads to their isolation, the loss of genetic diversity and, in the long term, their extinction (Frankham et al. 2010). Therefore, knowledge of the level and distribution of genetic diversity is necessary to determine the status of the remaining populations.
In this study, we aimed to estimate genetic variation and identify patterns of the population structure of E. europaeus in the north-eastern Iberian Peninsula. We also analysed some specimens from northwest Iberia for comparison. We asked if (i) the genetic diversity values were comparable between north-western and eastern populations, (ii) the genetic differences between these two major regions coincided with the phylogeographic distribution of lineages, and (iii) the hedgehogs of the studied regions retained genetic diversity and/or were genetically isolated. We paid particular attention to the Barcelona Zoo population, as it represents a vulnerable urban population with conservation priority. This information will permit us to clarify the connectivity of hedgehog populations in north-eastern Iberia, to know their conservation status, and to design conservation strategies for this species.
Material and methods
Samples
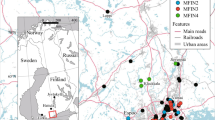
Ninety-two specimens of European hedgehogs, E. europaeus, from the northern region of the Iberian Peninsula (78 from the north-eastern region and 14 from the north-western region) were sampled from 2009 to 2015 (Fig. 1, Supporting Information, Table S1). This sampling included a natural population within Barcelona city, the members of which live freely within the zoo facilities, sampled in 2009. These hedgehogs can move freely in and out of the zoo, but this area represents an isolated green urban habitat in the city (Fig. 1). Hair follicles of the zoo animals and those from Park Agrari (Supporting Information, Table S1) were sampled during nocturnal sampling campaigns. The rest of the samples were obtained from individuals from animal rescue centres and from hedgehogs found dead along roads. We collected muscles from roadkill animals and from animals that died in rescue centres, and sampled hair follicles from live animals.
Map of sampling locations. Sample dots are coloured according to BAPS with rims referring to STRUCTURE clustering (black: Girona; green-grey: Central Catalonia; red: Zoo, see Fig. 2). Black solid lines represent populations defined for posterior analyses: Girona, Zoo and Central Catalonia (rest of samples). The grey solid line represents substructure within the Central Catalonia cluster identified by Geneland. Red dotted lines indicated principal routes (motorways). Black dotted lines represent “Massís de les Gavarres” and “Massís de Cadiretes” mountains. Map of the Barcelona city indicating the Zoo is also shown
Microsatellite genotyping
The total DNA was extracted from the muscle tissue using the Realpure Genomic DNA extraction toolkit (Durviz SL, Valencia, Spain) following the manufacturer’s instructions. For hair follicles, we added 1 M dithiothreitol (DTT) to proteinase K at the first step of the Durvitz protocol according to Köchl et al. (2005).
Eleven microsatellite loci, EEU1, EEU2, EEU3, EEU4, EEU5, EEU6 (Becher and Griffiths 1997), EEU12, EEU36, EEU37, EEU43 and EEU54 (Henderson et al. 2000), were amplified (Supporting Information, Table S2). The forward primers were end-labelled with fluorescent dye. The PCRs (20 μl) contained 2 μl DNA extraction, 2 μl Buffer (BIOLINE) 10 × , 0.1 μl of Taq DNA polymerase (BIOLINE) (5 u/μl), 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM), 2 μl dNTP MIX (2 mM) and 0.6 μl MgCl2 (50 mM). The thermal cycling conditions consisted of an initial denaturation step at 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 56–65 °C (Supporting Information, Table S2) for 1.5 min and 72 °C for 1.5 min, and a final step of 72 °C for 10 min. After amplification, the genotypes were visualized using an Applied Biosystem 3130 Genetic Analyser and interpreted visually with GeneMapper 4.0 software. To check for large allele dropouts and null alleles, the data were analysed using MICROCHECKER v 2.2.1 (Van Oosterhout et al. 2004) and FreeNA (Chapuis and Estoup 2007).
Data analysis
The minimum number of homogeneous units (K = 1–6) was estimated using the Bayesian Markov chain Monte Carlo approach method of STRUCTURE version 2.3.3 (Pritchard et al. 2000). Analyses were carried out with the admixture model and correlated allele frequencies without prior population information and with a burn-in period of 200,000 steps followed by 1,000,000 Monte Carlo replicates. Twenty independent runs were conducted for each K value tested. The optimal K value was selected following the recommendations of Pritchard et al. (2000) and according to the method by Evanno et al. (2005), implemented in STRUCTURE Harvester (Earl and vonHoldt 2012). In addition, the spatial Bayesian clustering methods implemented in BAPS v. 6.0 (Corander and Marttinen 2006) and GENELAND v.4.9.2 (Guillot et al. 2005) were used to identify spatially clustered individuals.
Because the above Bayesian methods are highly sensitive to uneven sample sizes among populations (Puechmaille 2016), we also used discriminant analysis of principal components (DAPC) (Jombart et al. 2010) to determine the number of genetic groups using the package Adegenet 2.1.3 (Jombart 2008) implemented in R 4.0.3. (R Core Team 2020). This method is free of classical assumptions on Hardy–Weinberg and linkage equilibrium and performs better than Bayesian methods in obtaining complex population structures (Jombart 2008). The a-score index was used to find the optimal number of principal components (PCs) that should be retained in the DAPC analysis to avoid overfitting of the model.
Finally, a Mantel test implemented in GENEALEX v 6.5 (Peakall and Smouse 2012) was used to test the correlations of the geographic and genetic distances (isolation by distance, IBD) in all the studied areas, and in the north-western and north-eastern regions individually, and the significance of the correlations was estimated by 999 random permutations.
Once homogeneous units were identified, genetic diversity parameters, number of alleles (A) and expected heterozygosity (Hs), were estimated for each cluster using the program FSTAT 2.9.3 (Goudet 2001). We used the software HR-RARE (Kalinowski 2005) to estimate the allelic richness (Ar) and the private allelic richness (PAr) in each cluster, by using the rarefaction method that standardize the number of alleles to the smallest population sample in our data set. Rarefaction was also used to compare Ar and PAr values per region (northeast and northwest) standardizing the number of populations sampled per region. Genotypic distributions at each cluster were tested for conformance to Hardy–Weinberg expectations by the exact test implemented in Genepop 4.0 (Raymond and Rousset 1995). The unbiased composite-linkage disequilibrium measure (D) of this program tested possible gametic disequilibria for all possible pairs of loci in each cluster. The sequential Benjamini-Yekutieli correction (Narum 2006) was applied to every single test to adjust the significance levels for multiple simultaneous comparisons. Allele frequency differences among clusters were evaluated by the exact probability test of Genepop 4.0. The weighted analyses of variance in the allele frequencies (pairwise FST), was used to assess genetic differentiation between each pair of clusters, and their significance were tested by randomizing genotypes between cluster pairs by performing 1000 permutations with FSTAT 2.9.3. It is expected that if gene flow exists among clusters we will find high genetic diversity values associated with a low genetic differentiation (FST). Contemporary migration rates (M) between cluster pairs were estimated using Bayesas 1.3 (Wilson and Rannala 2003). We performed 3 × 106 iterations, and data were recorded by 2000 iterations, with a burn-in period of 106 iterations, to ensure that the starting parameters of the model were randomized. The delta values were adjusted following the recommendations in the user manual. We performed five runs using different starting points and kept the results for the highest likelihood model (Faubet et al. 2007). The analysis of molecular variance (AMOVA) implemented in ARLEQUIN 3.1 (Schneider et al. 2000) quantified the percentage of spatial components on the total genetic variance (FST) according to a hierarchical model.
The Bottleneck program (Piry et al. 1999) was used to detect evidence of recent population bottlenecks in the studied populations. We used a two-phased model of mutation (TMP) with default values (70% single-step mutation and 30% multiple-step mutation).
Although our sampling was expected to mostly include unrelated individuals, some highly related individuals could not be discarded in the Barcelona Zoo population. Therefore, we used COLONY v2.0.6.6 (Jones and Wang 2010) to allocate individuals into sibling groups of full-sib families nested within half-sib families without information on parental genotypes. We tested COLONY forcing polyandry (Morán et al. 2009) and assumed an error rate of 2% caused by allelic dropouts and 0.1% erroneous allele sizing. Afterwards, we tested whether genetic differentiation was maintained when highly closely related individuals (full-sibs) were excluded from the analysis.
Results
We observed an average of 2.1% missing values, ranging from 0% at the EEU5 locus to 9.7% at the EEU12 locus. Missing values were confirmed after reanalyses, and genotyping for each locus was confirmed by the reanalysis of 50% of individuals. The number of alleles per locus ranged between 4 (EEU36) and 19 (EEU43), with an average number of alleles of A = 5.841. While the gene diversity (HS = 0.604) was high overall, it was lower in the north-eastern region (HS = 0.573) than in the north-western populations (HS = 0.696). Allelic richness standardized by the number of populations sampled per region was also higher in the northwest than in the northeast (4.964 vs 3.925, Table 1), and a Student’s t-test indicated that both Ar and PAr values differed significantly between these two regions (0.001 < P < 0.05 in both cases) (Table 1).
Distribution of genetic diversity and identification of clusters
At the uppermost level of the hierarchy, STRUCTURE clearly differentiated samples from north-western and north-eastern Iberia (K = 2). Individuals in the north-western region had the highest assigned coefficients (> 0.95), whereas some individuals from the northeast showed admixed ancestry of the two clusters (Fig. 2A.1, Supporting Information, Table S1). STRUCTURE did not detect substructure within the north-western cluster (K = 1), but four different groups were identified within the north-eastern region (K = 4, Fig. 2A.2). Individuals from the most north-eastern area (Girona) and those from the Zoo population were clearly grouped into two different clusters, with some level of admixture in a few individuals. The two remaining clusters were distributed among individuals from Central Catalonia with only a slight pattern of distribution. The results of Bayesian analyses considering the spatial location of individuals mostly agreed with the STRUCTURE results. BAPS and GENELAND identified two clusters that coincided with those identified by STRUCTURE in Girona and Zoo. BAPS grouped all remaining individuals in a single third cluster (Central Catalonia, Fig. 2B), whereas GENELAND clearly grouped these individuals into two clusters that separated individuals from the Barcelona area within the Central Catalonia cluster (Fig. 2C, Supporting Information, Table S1).
Admixture analyses by STRUCTURE, BAPS and GENELAND. Each individual is represented as a vertical bar partitioned into segments according to the proportion of the genome belonging to each of the identified clusters (K). A STRUCTURE: A.1: all samples (K = 2), A.2: north-eastern samples (K = 4). B BAPS with north-eastern samples (K = 3). C GENELAND with north-eastern samples (K = 4)
A model of four populations among all individuals was also estimated by the DAPC analyses. According to a-score optimization, we retained 11 principal components (PCs) and two discriminant functions (DA) that explained 92.5% of the variance in the data. The DAPC scatterplot on the two first DA grouped individuals into four clusters (Girona, Central Catalonia, Zoo and Northwest) (Fig. 3); which mainly coincided with the Bayesian clustering. The first discriminant axis of the DAPC explained 76.2% of the total variation and differentiated the northeast and northwest groups. The second axis described 16.3% of the variation and distinguished samples from the three north-eastern populations despite some individuals from Girona and Central Catalonia showed an overlapped distribution. At the individual level, discordances in the assignments were observed only in a few individuals assigned to Girona by the DAPC and to Central Catalonia by Bayesian methods. At a micro-geographical scale, the DAPC analyses did not detect any level of substructure within clusters except within the Central Catalonia group. For this subset, DAPC analyses retained 5 PCs and a single discriminant function that explained 100% of the variation. This substructure grouped individuals in two clusters that differentiated individuals from the area of Barcelona from the rest of Central Catalonia and more or less coincided with the clustering described by GENELAND (Fig. 3).
DAPC scatterplot. A Clustering of all individuals coloured according to the genetic cluster obtained by K-means clustering and assignment of individuals to each cluster. B DAPC sub-clustering and individual assignments within the Central Catalonia cluster, where a single DA explained genetic variation
Taking into account the most coincident results between all the Bayesian and the DAPC analyses, we considered three populations in the north-eastern region (Girona, Central Catalonia and Zoo) and a single fourth population in the north-western region in all posterior analyses.
Genetic diversity within clusters
After Benjamini-Yekuteli corrections, Hardy–Weinberg disequilibrium occurred in all but the northwestern clusters. In Girona and Central Catalonia, this was explained by disequilibrium at one (EEU1) and three loci (EEU5, EEU43 and EEU54), respectively. In the Zoo population, three (loci EEU1, EEU43 and EEU54) out of the 11 loci showed Hardy–Weinberg disequilibrium associated with a deficit of heterozygotes with index fixation (F) values of approximately 0.5 in all cases. MICRO-CHECKER analyses indicated that some of these deviations could be explained by the presence of null alleles at these loci. Null alleles have been previously described at the EEU43 and EEU54 loci (Zolotareva et al. 2021). The frequency of null alleles estimated by FreeNA (Chapuis and Estoup 2007) was higher than 0.2 in only two cases (EEU1 in Girona and EEU43 in Central Catalonia, Supporting Information, Table S3). According to results from FreeNA, estimation of the genetic differentiation (FST) excluding null alleles or using the ENA correction did not differ significantly, neither at the global level (FST = 0.129 versus FST = 0.123) nor in pairwise comparisons (Supporting Information, Table S3). Gametic disequilibrium was observed in the Central Catalonia and Zoo groups (in six and eight out of the 55 tests, respectively). In Central Catalonia, these groups comprised different loci, but in the Zoo, they involved mainly the EEU1 and EE54 loci. However, after Benjamini-Yekuteli corrections, only two significant deviations were detected in Central Catalonia (EEU6–EEU43; EEU2–EEU3 loci) and one in the Zoo (EEU3–EEU12 loci).
The highest values of genetic diversity were found in the north-western group (Ar = 4.964; PAr = 3.427, Hs = 0.696), and the lowest was found in Girona (Ar = 3.116; PAr = 1.578, Hs = 0.541). For all loci, the presence of private alleles ranged from 31 in the northwest cluster to 1 in Girona. Values of genetic diversity differed significantly among samples only when the north-western cluster was included in comparisons.
The Garza and Williamson (2001) index value, calculated by ARLEQUIN, was approximately 0.3–0.4, indicating a recent reduction in population size in all clusters. However, no signals of recent bottlenecks were detected by testing heterozygote excess in the BOTTLENECK program.
The study of the family relationships indicated no full-sib families within any cluster except in the Zoo population, where a single full-sib family of 4 members was identified. Genetic differentiation estimated retaining only one individual from this full-sib family did not differ significantly from values with all data (FST = 0.129 versus FST = 0.124).
Population structure
Hierarchical analyses of molecular variance (AMOVA) assigned a higher genetic variance to the differences between north-western and north-eastern clusters (11.56%) than among groups within these clusters (6.99%), with 81.43% of variance assigned within populations. All components of variance contributed significantly to genetic differentiation (P < 0.001). Overall FST was 0.129, but it reduced to 0.087 when the north-western cluster was excluded. Significant FST values were observed between all cluster pairs (P < 0.05). The highest FST values were observed between the north-western and north-eastern groups. High differentiation was also found between the Girona cluster and Zoo population, and the lowest FST was between Zoo and Central Catalonia clusters (Table 2).
In the north-eastern region, the contemporary migration rates (m) between cluster pairs, measured by Bayesass, were mainly unidirectional. High migration rates were detected from Central Catalonia to the rest of the clusters (m = 0.29 in both cases), but these rates were low in the opposite direction (m < 0.01). However, these results should be interpreted with caution because the number of individuals in the Central Catalonia group doubled those in the Girona and Zoo groups, and recent gene flow can be overestimated with small sample sizes (Faubet et al. 2007). The lowest migration value was obtained from the Zoo to the Central Catalonia group (0.0093), contrasting with the migration rate calculated in the opposite direction.
The Mantel test considering individuals from all populations indicated a weak but significant pattern of IBD in all cases (R = 0.455; P = 0.01). However, all these correlations (even those that were significant) decreased when IBD was tested within the two regions, the north-western region (R = 0.232; P = 0.013) and the north-eastern region (R = 0.160; P = 0.010), separately.
Discussion
Population structure
The hedgehogs in the study area were clearly grouped into two genetically well-differentiated clusters: the north-western and the north-eastern clusters. These groups fully coincide with the distribution of the two pre-glacial phylogenetic lineages described in the Iberian Peninsula by mtDNA, and it seems that phylogeographic events could explain such differentiation (Sanz et al. 2021). Large genetic differentiation between populations of the eastern and western Peninsula has been observed in other small mammals, such as the Stone Marten Martes foina E. (Vergara et al. 2015). At a macrogeographical scale, the population structure of E. europaeus has also been related to the postglacial colonization of two different phylogeographic lineages in Eastern Europe (Zolotareva et al. 2021).
At a low hierarchical level, a significant genetic structure was also detected within the north-eastern group. Genetic differentiation has been described in small mammal species with territorial behaviours (Becher and Griffiths 1998). However, European hedgehogs are not territorial (Reeve 1982). The differentiation among clusters could then be related to isolation because of habitat fragmentation by natural or artificial barriers. In the north-eastern region, the distribution of three different populations was slightly related to an IBD pattern associated with limited dispersion. This weak IBD pattern was evidenced by the high admixture ancestry in some individuals, probably due to recent migrations from the Central Catalonia group to Girona and Zoo populations. Braaker et al. (2017) related a similar differentiation pattern in Zurich to major rivers and transportation axes that restricted gene flow. Alternatively, Osaka et al. (2021) failed to detect genetic differentiation among urban hedgehogs around Helsinki, but they suggested a lack of structure with an overall genetic homogeneity due to a recent natural expansion or an artificial introduction of the species, and not the absence of geographical barriers to gene flow. Additionally, Bolfiková et al. (2013) found four genetic hedgehog populations with high proportions of admixed individuals in a comparable territory in New Zealand (268,021 km2), but these authors studied introduced populations, and their diversity and population structure could be modelled by their introduction history.
Despite some gene flow among populations, the presence of a great number of private alleles (Table 1) and the significant genetic differentiation among clusters indicated some isolation, mainly in the Girona and Zoo populations. In addition, incipient population fragmentation was observed within the Central Catalonia cluster, partially isolating hedgehogs from the Barcelona area and possibly being responsible for the Hardy–Weinberg and gametic disequilibria found in this cluster. The genetic differences observed in north-eastern Iberia are comparable with those detected in other hedgehogs from urban areas, such as Oxfordshire (Becher and Griffiths 1998), Zurich (Braaker et al. 2017) or Berlin (Barthel et al. 2020), and in rural Danish populations (Rasmussen et al. 2020), all of which are associated with habitat fragmentation. The effect of natural landscape elements such as large rivers on the population structure pattern has been well described in hedgehogs by Braaker et al. (2017) and Ploi et al. (2020). In our region, the mountains of “Massís de les Gavarres” and the “Massís de Cadiretes” (altitude 533 and 519 m, respectively) could act as geographical barriers that partially limit gene flow between Girona and Central Catalonia. The presence of mountains as a physical barrier has been reported in other small mammals of the order Eulipotyphla, such as Sorex araneus L. (Moska et al. 2012), although in this case, the mountains are approximately 1000 m in elevation. The network of roads, mainly motorways, that are present in the studied area could also represent artificial barriers to the mobility of hedgehogs because only a few individuals are able to cross wide roads (Huijser and Bergers 2000; Rondinini and Doncaster 2002; Braaker et al. 2017). However, the admixture levels detected in Central Catalonia, which is an area crossed and surrounded by motorways (Fig. 1), are more concordant with the idea that hedgehogs crossroads regularly at night when human activity levels decrease (Dowding et al. 2010) and that hedgehogs use road verges as movement corridors (Doncaster et al. 2001). In this sense, Rasmussen et al. (2020) also failed to associate road density with hedgehog habitat fragmentation.
On the other hand, the Zoo group could be particularly isolated, with only some sporadic contact with urban hedgehogs from Barcelona (Central Catalonia cluster). According to the unidirectional gene flow observed, it seems that some individuals from Barcelona reach the zoo, but practically no hedgehog leaves. Zoo fences do not prevent movement in either direction, therefore the isolation of this group could be explained because hedgehogs find favourable habitats and resources within the zoo facilities, and they do not migrate to find better conditions (Doncaster et al. 2001). The Zoo could thus act as an ecological trap, a situation that has been described for hedgehogs and other mammal species (Bolfíková and Hulva 2012; Vlaschenko et al. 2019). Ecological trapping in isolated favourable habitat patches has been proposed to explain interspecific coexistence of E. europaeus and E. roumanicus in Central Europe (Bolfíková and Hulva 2012). In the absence of connectivity, in urban environments these ecological traps can lead to fatal consequences for the contained populations (Vlaschenko et al. 2019). The situation of the Zoo population is similar to that described in hedgehogs inhabiting green patches in Berlin. These habitats are fenced or surrounded by big streets and heavy-traffic roads. In this case, the restriction of gene flow leads to a population structure of gamodemes explained by the concentration of related individuals within these patches (Barthel et al. 2020).
Genetic diversity: north-western vs north-eastern populations
The genetic diversity values obtained in the north-western region of our study area are comparable with those observed in other European hedgehog populations (Hs = 0.70, Becher and Griffiths 1998; Hs = 0.69, Bolfíková and Hulva 2012). However, these studies included more extensive areas and higher numbers of individuals. Surprisingly, with only 14 sampled individuals, northwest Iberia presented the highest genetic diversity values, even when diversity estimates were corrected by the differences in sampling effort between the two regions. Similarly, mitochondrial analysis indicated high haplotype diversity and demographic stability in the northwest (Sanz et al. 2021). High gene diversity values in populations of E. europaeus could be a consequence of the multiple paternities of hedgehogs (Morán et al. 2009). Instead, the genetic diversity measured in the north-western populations contrasted with the lower values observed in the north-eastern group. According to mtDNA studies, this lower genetic diversity could be related to the phylogeographic isolation of the eastern region, which is surrounded by the Pyrenees Mountains and Ebro River. Alternatively, these results could also suggest a better conservation of hedgehog populations in the northwest, which could be explained by the low human population and road network densities. Genetic diversity values similar to those found in the north-eastern region have been described in urban hedgehogs from highly fragmented habitats in Switzerland (Braaker et al. 2017) and Oxfordshire (Becher and Griffiths 1998) and even in rural mosaiced habitats in Italy (Iannucci et al. 2019) and Central Europe (Bolfíková and Hulva 2012). Even lower values of genetic diversity have been found by Osaka et al. (2021) in Helsinki populations because of founder events during the recent expansion of the species. Additionally, genetic diversity values observed in north-eastern Iberian hedgehogs are clearly lower than those reported for other small mammals from the order Eulipotyphla, such as Sorex araneus in Europe (Andersson et al. 2004; Moska et al. 2012).
Conclusions and conservation perspectives
It is clear from our results that the vulnerable situation described above for Iberian hedgehogs is most evident in the natural population from the Barcelona Zoo. In fact, it was only in this population that we to a full-sib family of four members and Hardy–Weinberg disequilibrium associated with a heterozygote deficit that could indicate some level of inbreeding in this population. It is said that the Barcelona Zoo hedgehog population predates the existence of the zoo itself, and this was there when King Felipe V demolished a third of the city of Barcelona to build a military citadel. Since 1714, these hedgehogs have resisted military life, the construction of the zoo and the Spanish civil war, among other events that disturbed their wildlife. However, the high genetic isolation associated with inbreeding evidenced by microsatellite data from 2009 indicates that the short-term survival of this population may be imperilled. In fact, the last census of this population in 2018 considered the Zoo population to be almost extinct. This extremely vulnerable situation resembles that described in the Regent’s Park in London: another small fluctuating population of very few hedgehogs that breed in an isolated green area next to the Zoo, in the middle of a big city (https://www.royalparks.org.uk/managing-the-parks/conservation-and-improvement-projects/hedgehogs). In the Barcelona Zoo, since 2018, a programme based on the introduction of individuals rescued in wildlife rehabilitation centres has been set up to reinforce the population and counteract inbreeding.
Hedgehogs are animals that are regularly brought to wildlife rescue centres, and sometimes the areas from which these animals are accepted are not taken into account when they are released. Human-mediated translocation can occur between different locations and promote gene flow between natural fragmented habitats (Ploi et al. 2020; Rasmussen et al. 2020; Osaka et al. 2021). Translocation of rescued individuals has been proposed to explain the lack of genetic differentiation among demes of urban Berlin (Barthel et al. 2020). These practices are also common in our region, and might be diluting the genetic differences among locations. This could also explain why some individuals found in the same geographical locations belong to different genetic clusters. Therefore, natural hedgehog dispersal could be more reduced that we inferred, and fragmentation could be even more exacerbated. Whether these human-mediated translocations serve to avoid inbreeding and fragmentation or dilute the native population structure pattern might be questionable. Our preliminary results suggest that these practices could be used to reinforce endangered populations similar to those from the Barcelona Zoo, but genetic monitoring of these introductions to confirm the resulting increase in gene diversity is necessary. On the other hand, human-mediated translocations between areas that are fragmented by natural barriers should be avoided, as they might disrupt local adaptations (De Jong et al. 2020). Moreover, because regional differences in endo-parasites have been observed among different hedgehog populations in Denmark, the prevention of spreading infectious diseases is another argument for avoiding human-mediated translocation (Rasmussen et al. 2021).
Finally, the conservation of populations such as those from the Barcelona Zoo that persist in urban landscapes should be of special interest because of their special role in biodiversity conservation (Aronson et al. 2017). Green areas provide sufficient connectivity to avoid habitat fragmentation in the urban landscape of Berlin (Barthel et al. 2020), and, for some mammalian species, management strategies that take into account small urban gardens as ‘natural’ corridors have been used successfully (Van Helden et al. 2020). A similar management strategy could be promoted in Barcelona city for hedgehog conservation. On a larger geographical scale, monitoring hedgehog populations and actions to improve hedgehog habitats and natural corridors are also recommended in the entire north-eastern Iberian region.
References
Allendorf FW (1983) Isolation, gene flow, and genetic differentiation among populations. Genet Conserv 18:51–65
Andersson AC, Narain Y, Tegelström H, Fredga K (2004) No apparent reduction of gene flow in a hybrid zone between the West and North European karyotipic groups of the common shrew, Sorex araneus. Mol Ecol 13:1205–1215. https://doi.org/10.1111/j.1365-294X.2004.02146.x
Aronson MFJ, Lepczyk CA, Evans KL, Goddard MA, Lerman SB, MacIvor JS, Nilon CH, Vargo T (2017) Biodiversity in the city: key challenges for urban green space management. Front Ecol Environ 15(4):189–196. https://doi.org/10.1002/fee.1480
Bannikova AA, Lebedev VS, Abramov AV, Rozhnov VV (2014) Constrasting evolutionary history of hedgehogs and gymnures (Mammalia: Erinaceomorpha) as inferred from a multigene study. Biol J Linn Soc 112:499–519. https://doi.org/10.1111/bij.12299
Barthel LMF, Wehner D, Schmidt A et al (2020) Unexpected gene-flow in urban environments: The example of the european hedgehog. Animals 10:1–17. https://doi.org/10.3390/ani10122315
Becher SA, Griffiths R (1997) Isolation and characterization of six polymorphic microsatellite loci in the European hedgehog Erinaceus europaeus. Mol Ecol 6:89–90. https://doi.org/10.1046/j.1365-294x.1997.00159.x
Becher SA, Griffiths R (1998) Genetic differentiation among local populations of the European hedgehog (Erinaceus europaeus) in mosaic habitats. Mol Ecol 7:1599–1604. https://doi.org/10.1046/j.1365-294x.1998.00457.x
Bolfíková B, Hulva P (2012) Microevolution of sympatry: landscape genetics of hedgehogs Erinaceus europaeus and E. roumanicus in Central Europe. Heredity 108:248–255. https://doi.org/10.1038/hdy.2011.67
Bolfíková B, Konecný A, Pfäffle M, Skuballa J, Hulva P (2013) Population biology of establishment in New Zealand hedgehogs inferred from genetic and historical data: conflict or compromise? Mol Ecol 22:3709–3720. https://doi.org/10.1111/mec.12331
Braaker S, Kormann U, Bontadina F, Obrist MK (2017) Prediction of genetic connectivity in urban ecosystems by combining detailed movement data, genetic data and multi-path modelling. Landsc Urban Plan 160:107–114. https://doi.org/10.1016/j.landurbplan.2016.12.011
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631. https://doi.org/10.1016/j.landurbplan.2016.12.011
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843. https://doi.org/10.1111/j.1365-294X.2006.02994.x
De Jong JF, van Hooft P, Megens HJ et al (2020) Fragmentation and translocation distort the genetic landscape of ungulates: red deer in the Netherlands. Front Ecol Evol 8:1–12. https://doi.org/10.3389/fevo.2020.535715
De Poel JLV, Dekker J, Van Langevelde F (2015) Dutch hedgehogs Erinaceus europaeus are nowadays mainly found in urban areas, possibly due to the negative effects of badgers Meles meles. Wildlife Biol 21:51–55. https://doi.org/10.2981/wlb.00072
Doncaster CP (1992) Testing the role of intraguild predation in regulating hedgehog populations. Proc R Soc B Biol Sci 249:113–117. https://doi.org/10.1098/rspb.1992.0092
Doncaster CP, Rondinini C, Johnson PCD (2001) Field test for environmental correlates of dispersal in hedgehogs Erinaceus europaeus. J Anim Ecol 70:33–46. https://doi.org/10.1111/j.1365-2656.2001.00471.x
Dowding CV, Harris S, Poulton S, Baker PJ (2010) Nocturnal ranging behaviour of urban hedgehogs, Erinaceus europaeus, in relation to risk and reward. Anim Behav 80:13–21. https://doi.org/10.1016/j.anbehav.2010.04.007
Earl DA, vonHoldt BM (2012) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Faubet P, Waples RS, Gaggiotti OE (2007) Evaluating the performance of a multilocus Bayesian method for the estimation of migration rate. Mol Ecol 16:1149–1166. https://doi.org/10.1111/j.1365-294X.2007.03218.x
Forman RTT, Alexander LE (1998) Roads and their major ecological effects. Annu Rev Ecol Syst 29:207–231. https://doi.org/10.1146/annurev.ecolsys.29.1.207
Frankham R, Briscoe DA, Ballou JD (2010) Introduction to conservation genetics, 2nd edn. Cambridge University Press, UK
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318. https://doi.org/10.1046/j.1365-294x.2001.01190.x
Goudet J (2001) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486. https://doi.org/10.1093/oxfordjournals.jhered.a111627
Guillot G, Mortier F, Estoup A (2005) Geneland: a computer package for landscape genetics. Mol Ecol Notes 5:712–715. https://doi.org/10.1111/j.1471-8286.2005.01031.x
He K, Chen JH, Gould GC, Yamaguchi N, Ai HS, Wang YX, Zhang YP, Jiang XL (2012) An estimation of Erinaceidae Phylogeny. A combined analysis approach. PLoS ONE 7:1–14. https://doi.org/10.1371/journal.pone.0039304
Henderson M, Becher SA, Doncaster CP, Maclean N (2000) Five new polymorphic microsatellite loci in the European hedgehog Erinaceus europaeus. Mol Ecol 9:179–180. https://doi.org/10.1046/j.1365-294x.2000.01098-18.x
Hof AR, Bright PW (2009) The value of green-spaces in built-up areas for—western hedgehogs. Lutra 52:69–82
Hof AR, Bright PW (2010) The value of agri-environment schemes for macro-invertebrate feeders: hedgehogs on arable farms in Britain. Anim Conserv 13:467–473. https://doi.org/10.1111/j.1469-1795.2010.00359.x
Hof AR, Bright PW (2016) Quantifying the long-term decline of the West European hedgehog in England by subsampling citizen-science datasets. Eur J Wildl Res 62:407–413. https://doi.org/10.1007/s10344-016-1013-1
Huijser MP, Bergers PJM (2000) The effects of roads and traffic on hedgehog (Erinaceus europaeus) populations. Biol Conserv 95:111–116. https://doi.org/10.1016/S0006-3207(00)00006-9
Iannucci A, Baccetti N, Giannini F, Gotti C, Baratti M (2019) A genetic analysis of the European hedgehog (Erinaceus europaeus): an applicative case study to support its eradiation from Pianosa Island (Tuscan Archipielago). Conserv Gen 20:395–402. https://doi.org/10.1007/s10592-018-1078-3
IUCN (2021) The IUCN Red List of Threatened Species. Version 2021–2. https://www.iucnredlist.org. Downloaded on 01 December 2021
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devilland S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jones OR, Wang J (2010) COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 10:551–555. https://doi.org/10.1111/j.1755-0998.2009.02787.x
Kalinowski ST (2005) HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x
Köchl S, Niederstätter H, Parson W (2005) DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR. Methods Mol Biol 297:13–30. https://doi.org/10.1385/1-59259-867-6:013
Mathews F, Harrower C (2020) IUCN—compliant Red List for Britain’s Terrestrial Mammals. Assessment by the mammal society under contract to Natural England Natural Resources Wales and Scottish Natural Heritage. Natural England, Peterborough (ISBN 978-1-78354-485-1)
Micol T, Doncaster CP, Mackinlay LA (1994) Correlates of local variation in the abundance of hedgehogs Erinaceus europaeus. J Anim Ecol 63:851–860. https://doi.org/10.2307/5262
Morán S, Turner PD, Reilly CO (2009) Multiple paternity in the European hedgehog. J Zool 278:349–353. https://doi.org/10.1111/j.1469-7998.2009.00583.x
Morris PA (1988) A study of home range and movements in the hedgehog (Erinaceus europaeus). J Zool 214:433–449
Moska M, Wierzbicki H, Strzala T, Mucha A, Dobosz T (2012) Genetic structuring of the common shrew, Sorex araneus (Soricomorpha: Soricidae) in the Polish Sudetes may suggest ways of northwards colonization. Hereditas 149:197–206. https://doi.org/10.1111/j.1601-5223.2012.02266.x
Narum SR (2006) Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv Genet 7:783–787
Orlowski G, Nowak L (2004) Road mortality of hedgehogs Erinaceus spp. in farmland in lower Silesia (South-western Poland). Pol J Ecol 52:377–382
Osaka M, Pynnönen-Oudman K, Lavikainen A et al (2021) Genetic diversity and phylogeography of urban hedgehogs (Erinaceus europaeus) around Helsinki, Finland, revealed by mitochondrial DNA and microsatellite analyses. Mammal Res. https://doi.org/10.1007/s13364-021-00603-5
Palomo LJ, Gisbert J, Blanco JC (eds) (2007) Atlas y libro rojo de los mamíferos terrestres de España. Ministerio de Medio Ambiente, Madrid (Spain)
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Pettett CE, Moorhouse TP, Johnson PJ, Macdonald DW (2017) Factors affecting hedgehogs (Erinaceus europaeus) attraction to rural villages in arable landscapes. Eur J Wildl Res 63:54. https://doi.org/10.1007/s10344-017-1113-6
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503. https://doi.org/10.1093/jhered/90.4.502
Ploi K, Curto M, Bolfíková BC, Loudová M, Hulva P, Seiter A, Fuhrmann M, Winter S, Meimberg H (2020) Evaluating the impact of wildlife shelter management on the genetic diversity of Erinaceus europaeus and E. roumanicus in their contact zone. Animals 10:1452. https://doi.org/10.3390/ani10091452
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. https://doi.org/10.1111/1755-0998.12512
R Development Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. URL. https://www.R-project.org/
Rasmussen SL, Berg TB, Dabelsteen T, Jones OR (2019a) The ecology of suburban juvenile European hedgehogs (Erinaceus europaeus) in Denmark. Ecol Evol 9:13174–13187. https://doi.org/10.1002/ece3.5764
Rasmussen SL, Yashiro E, Sverrisdóttir E et al (2019b) Applying the GBS technique for the genomic characterization of a Danish population of European hedgehogs (Erinaceus europaeus). Genet Biodivers 3:78–86
Rasmussen SL, Nielsen JL, Jones OR, Berg TB, Pertoldi C (2020) Genetic structure of the European hedgehog (Erinaceus europaeus) in Denmark. PLosOne 15:e0227205. https://doi.org/10.1371/journal.pone.0227205
Rasmussen SL, Hallig J, van Wijk RE, Petersen HH (2021) An investigation of endoparasites and the determinants of parasite infection in European hedgehogs (Erinaceus europaeus) from Denmark. Int J Parasitol Parasites Wildl 16:217–227. https://doi.org/10.1016/j.ijppaw.2021.10.005
Raymond M, Rousset F (1995) GENEPOP: a population genetics software for exact test and ecumenicism. J Hered 86:248–249. https://doi.org/10.1093/oxfordjournals.jhered.a111573
Reeve NJ (1982) The home range of the hedgehogs as revealed by a radio tracking study. Symp Zool Soc London 49:207–230. https://doi.org/10.1111/j.1469-7998.1988.tb03751.x
Reeve NJ (1994) Hedgehogs. In: Poyser TAD (ed) Natural history. University of California, London
Rondinini C, Doncaster CP (2002) Roads as barriers to movement for hedgehogs. Funct Ecol 16:504–509. https://doi.org/10.1046/j.1365-2435.2002.00651.x
Santucci F, Emerson C, Hewitt GM (1998) Mitochondrial DNA phylogeography of European hedgehogs. Mol Ecol 7:1163–1172. https://doi.org/10.1046/j.1365-294x.1998.00436.x
Sanz N, Vidal O, García S, Molina R, Araguas RM (2021) Molecular characterization of spiny hedgehogs of the Iberian Peninsula: the missing link in the postglacial colonization of the western European hedgehog. Mamm Res 66:187–200. https://doi.org/10.1007/s13364-020-00550-7
Schneider S, Roessli D, Excoffier L (2000) Arlequin: a software for population genetics data analysis. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Switzerland
Seddon JM, Santucci F, Reeve NJ, Hewitt GM (2001) DNA footprints of European hedgehogs, Erinaceus europaeus and E. concolor.: Pleistocene refugia, postglacial expansion and colonization routes. Mol Ecol 10:2187–2198. https://doi.org/10.1046/j.0962-1083.2001.01357.x
Taucher AL, Gloor S, Dietrich A et al (2020) Decline in distribution and abundance: urban hedgehogs under pressure. Animals 10:1–22. https://doi.org/10.3390/ani10091606
Van Helden BE, Close PG, Steven R (2020) Mammal conservation in a changing world: can urban gardens play a role? Urban Ecosyst 23:555–567. https://doi.org/10.1007/s11252-020-00935-1
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICROCHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Vergara M, Basto MP, Madeira MJ, Gómez-Moliner BJ, Santos-Reis M, Fernandes C, Ruiz-González A (2015) Inferring population genetic structure in widely and continuously distributed carnivores: the stone marten Martes foina as a case study. Plos one 10:e0134257. https://doi.org/10.1371/journal.pone.0134257
Vlaschenko A, Kovalov V, Hukov V et al (2019) An example of ecological traps for bats in the urban environment. Eur J Wildl Res 65:1–5. https://doi.org/10.1007/s10344-019-1252-z
Williams BM, Baker PJ, Thomas E et al (2018) Reduced occupancy of hedgehogs (Erinaceus europaeus) in rural England and Wales: the influence of habitat and an asymmetric intra-guild predator. Sci Rep 8:17–20. https://doi.org/10.1038/s41598-018-30130-4
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Young RP, Davison J, Trewby ID, Wilson GJ, Delahay RJ, Doncaster CP (2006) Abundance of hedgehogs (Erinaceus europaeus) in relation to the density and distribution of badgers (Meles meles). J Zool 269:349–356. https://doi.org/10.1111/j.1469-7998.2006.00078.x
Zingg R (1994) Aktivität sowie Habitat- und Raumnutzung von Igeln (Erinaceus europaeus) in einem ländlichen Siedlungsgebiet. Ph.D. Thesis, Universität Zürich, Zurich, Swiss
Zolotareva KI, Belokon MM, Belokon YS, Rutovskaya MV, Hlyap LA, Sarykov VP, Politov DV, Lebedev VS, Bannikova AA (2021) Genetic diversity and structure of hedgehogs Erinaceus europaeus and Erinaceus roumanicus: evidence for ongoing hybridization in Eastern Europe. Biol J Linn Soc 132:174–195. https://doi.org/10.1093/biolinnean/blaa135
Acknowledgements
We thank to Xavier Puig, Enric Fàbregues, Gonzalo Alarcos, Javier Talegón, Laia Planella and Xisco Roig for samples of this research. We also thank the personal of the Zoo and the Wildlife rehabilitation Centers of Torreferrussa and Aiguamolls de L’Empordà its collaboration. We received financial support from the Barcelona Zoo Foundation. Cesca Rivas provided technical support. We appreciate valuable comments from two anonymous reviewers to improve the previous version of the manuscript.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Handling editor: Laura Iacolina.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Araguas, RM., Vidal, O., García, S. et al. Genetic diversity and population structure of the Western European hedgehog, Erinaceus europaeus: conservation status of populations in the Iberian Peninsula. Mamm Biol 102, 375–386 (2022). https://doi.org/10.1007/s42991-022-00235-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42991-022-00235-5