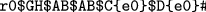Abstract
The bigraph theory, devised by Robin Milner, is a recent mathematical framework for concurrent processes. Its generality is able to subsume many existing process calculi, for example, CCS, CSP, and Petri nets. Further, it provides a uniform proof of bisimilarity, which is a congruence. We present the first canonical string encoding for pure and lean bigraphs by lifting the breadth-first canonical form of rooted unordered trees to a unique representation for bigraphs up to isomorphism (i.e., lean-support equivalence). The encoding’s applicability is limited to atomic alphabets. The time complexity is \(O(n^{2}k\, d \log {d})\), where n is the number of places, d the degree of the place graph and k the maximum arity of a bigraph’s signature. We provide proof of the correctness of our method and also conduct experimental measurements to assess the complexity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Graphs are well-understood and useful mathematical abstractions [20, 44]. Informally speaking, ordinary graphs comprise nodes and edges which allow representing “binary relations between nodes” [13, p. 463]. Moreover, graphs can be equipped with any non-trivially semantic meaning or structural extension, e.g., to represent Boolean functions by propositional directed acyclic graphs [45], or to describe static system structures of concurrent systems [20, p. 8]. An extension called hypergraphs [9] allows multiple links between nodes via hyperedges and lifts the binary relation limitation. Due to their general formalism, they found to be useful for object representation, and in this respect, are commonly used for modeling complex structured data [14] within a variety of different domains. To give one particular example, they are heavily employed in the field of software development as underpinning for models, where different structures on different levels are handled as graphs [20, pp. 7].
With this in mind, we often need to decide if two graphs are equal (i.e., isomorphic), which is a fundamental question in graph theory [26]. Here we can distinguish between two strands (cf. the taxonomy of matching problems in [18]). Sometimes a non-exact or approximate match is sufficient when we only need to check if a substructure is contained within another target graph. Roughly, given two graphs \(G = (V_G,E_G)\) and \(H=(V_H,E_H)\), the task is to find a subgraph of G which is isomorphic to H. This is referred to as subgraph isomorphism problem, which will not be treated here. An application of subgraph isomorphism might be, as mentioned in [43], to determine whether a certain chemical substance is present in a given compound. In the second case, we might be interested in an exact match. This particular field is entitled as exact graph matching or graph isomorphism, which is a subclass of the subgraph isomorphism problem. Specifically, two graphs \(F = (V_F, E_F)\) and \(H = (V_H, E_H)\) are isomorphic \(F \cong H\) if they are structurally the same graph (see [44, Def. 7.1]). For example, within a software application, we want to avoid re-creating the same object model (i.e., a graph) multiple times, meaning, we must check if the same structure already exists. Several possible matching solutions for different graph types can be identified in the literature, where we wish to address a few in “Related Work” later. For further reference, we wish to mention the survey of Conte et al. [18], where a comprehensive overview is presented of many applications of graphs and graph matching algorithms.
Contribution. In this paper, our approach is devoted to the bigraph isomorphism problem for the pure case. Essentially, a bigraph \(B = \langle B^P, B^L\rangle\) is a hierarchical graph comprising two independent graph structures: The place graph \(B^P\) describes the nesting of nodes in a tree-like structure, the connection among nodes is expressed by the link graph \(B^L\); both relying on the same node-set (see Def. 6). Our contribution is the computation of a unique representation of a bigraph up to isomorphism. The unique representation of a graph is also referred to as canonical form or canonical labeling in this paper. If we can find a canonical mapping, we can reduce the bigraph isomorphism problem to a string matching problem (i.e., checking the corresponding canonical labelings of two bigraphs on equality), without resorting to brute force. To do so, we must respect the (lean-) support equivalence property for two bigraphs (see [33, p. 16] and also Def. 8 and 9), which corresponds to finding a support translation \(\rho\) such that  (see Def. 8). Consequently, we can check for two bigraphs F and G whether their canonical forms are equal, rather than isomorphic (cf. also [38]). Given the computational complexity of an isomorphism test, “a canonization algorithm is often preferable to an isomorphism test” [38, p. 1]. (This is not to be confused with the well-defined bigraph matching in the literature, see “Related Work”.) As a result, the problem becomes O(n), where n is the length of the string encoding (precluding the processing time for computing the canonical form). This means that testing the equality of graphs is, therefore, easy, whereas the computation of the canonical labeling is often not [26]. However, it allows graphs to be treated separately instead of comparing graphs pairwise [38] (see also “Discussion”). Furthermore, existing software implementations such as nauty [30] prove that efficient isomorphism tests based on graph canonization are realizable. To the best of our knowledge, no such string encoding exists for pure and lean bigraphs currently.
(see Def. 8). Consequently, we can check for two bigraphs F and G whether their canonical forms are equal, rather than isomorphic (cf. also [38]). Given the computational complexity of an isomorphism test, “a canonization algorithm is often preferable to an isomorphism test” [38, p. 1]. (This is not to be confused with the well-defined bigraph matching in the literature, see “Related Work”.) As a result, the problem becomes O(n), where n is the length of the string encoding (precluding the processing time for computing the canonical form). This means that testing the equality of graphs is, therefore, easy, whereas the computation of the canonical labeling is often not [26]. However, it allows graphs to be treated separately instead of comparing graphs pairwise [38] (see also “Discussion”). Furthermore, existing software implementations such as nauty [30] prove that efficient isomorphism tests based on graph canonization are realizable. To the best of our knowledge, no such string encoding exists for pure and lean bigraphs currently.
Structure. Our paper is structured as follows. The necessary background about some elementary graph structures and the bigraphical concepts are provided in “Graph-Theoretical Background”, which are useful to observe how ordinary graphs relate to bigraphs. In “Bigraphical Canonical String Encoding”, we first provide the key ideas that are used in our approach, then present our implementation to compute the canonical string of a pure bigraph, provide sufficient examples and address the time complexity. In “Discussion”, the paper continues with a discussion, reviewing the problem of graph isomorphism in general, canonical labelings, and the one of bigraph matching, and reflects the practical application of our work. Finally, we conclude our paper with “Bigraphical Canonical String Encoding” by giving some final remarks.
Graph-Theoretical Background
In this section, we recall bigraphs and their underlying mathematical primitives, which are helpful for the understanding of the presentation of our canonical string encoding algorithm presented in “Bigraphical Canonical String Encoding”.
Elementary Graph Structures
Definition 1
(Tree) A connected graph with no cycles of the form \(G = (V_G, E_G)\) is termed unordered undirected tree, or just tree. Edges and vertices of G have no labels implying no special ordering, and edges also have no direction, meaning, \(e = (v_i,v_j)\) implies \(e = (v_j,v_i)\) for each edge \(e \in E_G\).
Definition 2
(Rooted tree) (after [42, p. 269]) A rooted tree is a triplet \(G = (V_G, E_G, r_G)\), where \((V_G, E_G)\) is a unrooted tree as by Def. 1, each \(v \in V_G\) has a distinct parent and \(r_G\) is some vertex in \(V_G\) which is called the root, implicitly specifying the parent-child-relationship, thus, the edge direction. Vertices under the same parent are called siblings, vertices with no children are called leaves of the tree.
Definition 3
(Hypergraph) (after [9, p. 226]) A hypergraph \(H = (V, E)\) contains a finite set of vertices V and a family of sets \(E = (e_i)_{i\in I}\) of non-empty subsets of V called the hyperedges or edges of H, where I is a finite index set.
Bigraphs
Here we provide the standard definition of concrete bigraphs given in [33] for the pure case. Bigraphs are not only a formal graphical model but also provide a graph and term representation (see [33]). An example of a bigraph \(B:\langle 2,\{x_1,x_2\}\rangle \rightarrow \langle 1, \{y_1,y_2\}\rangle\) is depicted in Fig. 1 and the formal definition of its algebraic graph representation is given with Def. 6. As a shorthand what follows, a finite ordinal n often reads \(n = \{0,1,\ldots ,n-1\}\). Further, \(A \uplus B\) is the disjoint union of sets A and B, and \({\mathcal {X}}\) is an infinite alphabet.
Definition 4
(Bigraph interface) (after [33, Def. 2.3]) An interface for bigraphs is a pair \(I = \langle m,X \rangle\) of a place graph interface and a link graph interface, where \(X \subset {\mathcal {X}}\) is a finite set of names and m is called the width of I. We call I nullary, unary, or multiary if m is 0, 1, or \(>1\), respectively.Footnote 1
A concrete bigraph (see Def. 6) is always defined over a signature \({\varSigma }\), which specifies the syntax of the bigraph:
Definition 5
T(Basic signature) (after [33, p. 7]) A basic signature \({\varSigma }\) takes the form \(({\mathcal {K}}, ar)\). It has a set \({\mathcal {K}}\) whose elements are kinds of node called controls, and a map \(ar:{\mathcal {K}} \rightarrow {\mathbb {N}}\) assigning an arity, a natural number, to each control. The signature is denoted by \({\mathcal {K}}\) when the arity is understood. A bigraph over \({\mathcal {K}}\) assigns to each node a control, whose arity indexes the ports of a node, where links may be connected.
We use sans-serif letters (A, B, ..., Z) for controls of \({\varSigma }\) to distinguish them easily from other symbols. For example, the bigraph in Fig. 1 is defined over the signature \({\varSigma }= \{\textsf {A}:0, \textsf {B}:0, \textsf {D}:2, \textsf {E}:2, \textsf {F}:1,\textsf {G}:0,\textsf {H}:0,\textsf {Q}:1,\textsf {R}:1\}\).
Definition 6
(Concrete bigraph) (after [33, Def. 2.3]) A concrete bigraph is a quintuplet
comprising a concrete place graph \(F^P =(V_F, ctrl_F,prnt_F):k \rightarrow m\) and a concrete link graph \(F^L = (V_F,E_F,ctrl_F,link_F):X \rightarrow Y\). A concrete bigraph is also written as \(F = \langle F^P, F^L\rangle\).
-
\(v \in V_F\) is a node of the node set of F that is shared among the place graph and the link graph.
-
\(e \in E_F\) is a hyperedge of the set of hyperedges of F.
-
\(ctrl_F:V_F \rightarrow {\mathcal {K}}\) is the control map. Each node is assigned a control from the signature \({\mathcal {K}}\).
-
\(prnt_{F}:k \uplus V_F \rightarrow V_F \uplus m\) is the parent map which defines the parent-child-relationship of the place graph’s nodes. Thus, expressing the locality of nodes.
-
\(link_{F}:X \uplus P_F \rightarrow E_F \uplus Y\) is the link map of the link graph to express connectivity among the nodes. The disjoint union
 is the set of ports of F.
is the set of ports of F.
In particular, the place graph of F is a forest, and the link graph of F is a hypergraph. Both structures are defined and constructed independently. This allows to model the two elementary aspects for global computing, namely, locality and interaction of processes, which are prominent for recent developments in this area [11].
Example of a bigraph \(B:\langle 2, \{x_1,x_2\} \rangle \rightarrow \langle 1,\{y_1,y_2\} \rangle\) in a slightly different graphical display as presented in [33]
Support of a bigraph. Now we come to the definition of the support equivalence of a bigraph, which declares an essential property for our canonical string encoding that has to be considered to be applied for the bigraph isomorphism problem.
Definition 7
(Support for bigraphs) (after [33, Def. 2.4]) To each place graph, link graph or bigraph F is assigned a finite set |F|, its support, For a place graph we define \(|F| = V_F\), and for a link graph or bigraph we define \(|F| = V_F \uplus E_F\).
Definition 8
(Support equivalence and support translation) (after [33, Def. 2.4]) Two bigraphs F and G in the same homset are said to be support equivalent, and we write \(F \bumpeq G\), if there is a support translation of F by \(\rho\) that uniquely determines G, meaning, the support translation \(\rho\) is a bijection \(\rho :|F| \rightarrow |G|\) that respects the structure of F such that  . By Def. 7, the support translation consists of a pair of bijections \(\rho_{V}\colon {\text{V}}_{{\text{F}}} \to {\text{V}}_{{\text{G}}}\) and \(\rho_{E}\colon {\text{E}}_{{\text{F}}} \to {\text{E}}_{{\text{G}}}\).
. By Def. 7, the support translation consists of a pair of bijections \(\rho_{V}\colon {\text{V}}_{{\text{F}}} \to {\text{V}}_{{\text{G}}}\) and \(\rho_{E}\colon {\text{E}}_{{\text{F}}} \to {\text{E}}_{{\text{G}}}\).
With regard to Def. 4, we write \((I \rightarrow J)\) for the same homset of I and J, meaning the set of bigraphs \(f:I \rightarrow J\) with the same interfaces (cf. [33, Def. 2.8]).
Definition 9
(Lean-support equivalence) (after [33, Def. 2.19]) A bigraph is lean if it has no idle edges. Two bigraphs F and G are lean-support equivalent, written \(F \Bumpeq G\), if they are support equivalent (Def. 8) ignoring their idle edges. Composition and tensor product preserve this equivalence.
Definition 10
(Bigraph isomorphism) Two bigraphs F and G are isomorphic if and only if they are support equivalent. By Def. 8, we write \(F \bumpeq G\). Similarly, two lean bigraphs F and G are isomorphic if and only if they are lean-support equivalent. By Def. 9, we write \(F \Bumpeq G\).
Bigraphical Canonical String Encoding
In this section, we introduce the underlying key idea behind our approach. The authors in [15] define two unique representation for ordered rooted trees: the breadth-first canonical form (BFCF) and depth-first canonical form (DFCF), where the former is the one we exploit. For understanding of our extension, we recall the bottom-up procedure of [15] that obtains the BFCF of a labeled rooted unordered tree, which relates to a place graph.
The tree is traversed level by level, starting from the bottom. At each level, families of siblings are reordered from small to large. This step is performed until the children of the root are reordered. By recursion all subtrees are in the correct form. Then, performing a breadth-first traversal, all labels are recorded from left to right, where families of siblings are partitioned by the symbol  and the end of the string is denoted by #. Lemma 2.1 in [15] states that for each labeled rooted ordered tree there exists a corresponding unique breadth-first string encoding, and vice versa. Based on this BFCF description, the breadth-first canonical string (BFCS) for rooted unordered trees is defined afterwards. For a rooted unordered tree, many derivatives of rooted ordered tree exist according to the node order and, therefore, many corresponding canonical forms can be deduced from them. The minimal string encoding according to the lexicographic order among all these encodings is the one to use for representing that rooted unordered tree (see [15, p. 207]).
and the end of the string is denoted by #. Lemma 2.1 in [15] states that for each labeled rooted ordered tree there exists a corresponding unique breadth-first string encoding, and vice versa. Based on this BFCF description, the breadth-first canonical string (BFCS) for rooted unordered trees is defined afterwards. For a rooted unordered tree, many derivatives of rooted ordered tree exist according to the node order and, therefore, many corresponding canonical forms can be deduced from them. The minimal string encoding according to the lexicographic order among all these encodings is the one to use for representing that rooted unordered tree (see [15, p. 207]).
Method
We are now ready to show the core of our method, namely, how to obtain a breadth-first string encoding (BFSE) of a pure bigraph. The cornerstone of our method is the uniqueness of the canonical form for a labeled rooted ordered tree which guarantees “the uniqueness of [...] the BFCF for a labeled rooted unordered tree” [15, p. 207]. To bring the BFCF in relation to bigraphs, the rooted unordered tree corresponds to the place graph whose nodes are by definition “labeled nodes” because of their controls (see Def. 6).
In the parlance of [15], we assume for the rest of the paper that (a) there exists a total ordering between each of the control labels and link names; (b) the set of controls \({\mathcal {K}}\) of the signature \({\varSigma }\) are drawn from an atomic alphabet (i.e., single-character labels), which additionally contains four special symbols not in the alphabet, namely,  and # which denote sibling partition and the end of the string, respectively, further { and } to group the node’s links; (c) the group
and # which denote sibling partition and the end of the string, respectively, further { and } to group the node’s links; (c) the group  sorts smaller than any other symbol, # sorts greater than
sorts smaller than any other symbol, # sorts greater than  , and both sort greater than any other symbol in the alphabet of control labels and
, and both sort greater than any other symbol in the alphabet of control labels and  .
.
The procedure’s skeleton for obtaining the BFSE is presented in Algorithm 2 where we wish to give an outline in the following. The bigraph’s structure is sequentially translated, level by level, into a canonically ordered form using a breadth-first search (BFS) with a bottom–up step beginning with the root at index 0, instead of starting from the bottom compared to the BFCF procedure described in [15].

For the breadth-first traversal, we utilize parts of the approach explained in [7, 8]. The authors present a hybrid approach combining the top–down and bottom–up BFS by switching them based on the growing or shrinking frontier size of the nodes being visited. It may dramatically reduce the edges to be visited when traversing the place graph. Notably due to this necessity, the bottom–up approach is more preferable than the top–down one in some situations, particularly, when a large number of nodes is present in the frontier, meaning when the node degree of the place graph is high (see [7]). We recall the single bottom–up step in Algorithm 1 for our BFS to make this paper self-contained. Based on the bottom–up BFS, the canonical labeling procedure is directly applied to the place graph, at the same time, we check the link graph to respect the structural dependency of both bigraph constituents.
Place encoding. We start with Line 11 in Algorithm 2 which records the place encoding. The nodes are ordered from left to right (given by the BFS), and from small to large. Meaning, in each level, we first group the nodes by their parents, then sort the entries according to the following precedence: their control labels (including the parent and its children), the number of children and the total sum of the port count of all children. However, we respect the sorting of former parents when moving into the next level. Following the BFS level by level, the control label of each node is then appended to the string. If a node is a site, its index is recorded instead of the label. (We assume that the site indices are not contained in the alphabet before.) Furthermore, we record for each node the links it is connected to. Therefore, its links are fenced within

recording only their rewritten identifiers (which we explain in the next paragraph). This step only records edges and outer names by checking \(link_F(v_i) \ne \emptyset , v_i \in V_F\) (see Def. 6). To partition families of siblings, we use  , and to denote the end of the place encoding, we add #. In addition, we record
, and to denote the end of the place encoding, we add #. In addition, we record  in the next level for a node that had no children before and only if in the next level at least one parent has further nodes. For example, consider the left subtree B under A in Fig. 1. According to the above defined order, its encoding is
in the next level for a node that had no children before and only if in the next level at least one parent has further nodes. For example, consider the left subtree B under A in Fig. 1. According to the above defined order, its encoding is  . Because both D and E have no children, two
. Because both D and E have no children, two  symbols are recorded first before GH, which are the child nodes of F.
symbols are recorded first before GH, which are the child nodes of F.
Notice that this procedure is called iteratively inside a for each loop for each tree of the place graph’s forest (Line 4 in Algorithm 2). Consequently, the place graph encoding for each tree is finalized by # at the end. Further note that the first argument of the function bottom–up-step in Algorithm 1 takes not all places of the bigraph but rather \(B\!\downharpoonright ^{m'}\), the tree rooted at \(m'\) (i.e., the current root) which is the set of nodes and sites as defined in [27, Def. 7.2.9].
Link encoding. Now we proceed on with Line 15 of Algorithm 2, which treats the connections among the inner names and links (i.e., edges and outer names) only. The following explanation is just a matter of specifying a suitable ordering among the connections expressed by the link graph. Therefore, we apply a complete label rewriting of all edges due to the new ordering of the place graph. In bigraph matching the identity of these labels do not affect the rewriting result. Inner names and outer names remain untouched and preserve their natural ordering (cf. Def. 8 and Def. 9). A correct and unique re-labeling of edges is achieved by always traversing the place graph first, and based upon this ordering these link names are rewritten and recorded. We generate constant symbols with the following characteristics: rewritten edge identifiers sort greater than inner names, and inner names sort greater than outer names. Edge identifiers are prefixed by e, where for each character an integer is appended which is continuously incremented for every new unvisited link, e.g., e2 means that a previously unvisited edge \(e_i \in E_B\) was rewritten after two other (already rewritten) edges were visited.
The rest of the possible link patterns are recorded at the end of the string in the following order: (1) idle inner names,Footnote 2 (2) connections from inner names to edges, (3) connections from inner names to outer names, and (4) idle outer names. According to the above sequence, each link pair is separated by the symbol  , finally closing the link encoding with an additional #. For instance,
, finally closing the link encoding with an additional #. For instance,  constitutes a valid link encoding w.r.t. node-free link graphs. Here, x0 and x1 are idle inner names and y0 is an idle outer name. The middle part tells us that the inner name x2 is connected to the edge e3, and x3 is linked to the outer name y1.
constitutes a valid link encoding w.r.t. node-free link graphs. Here, x0 and x1 are idle inner names and y0 is an idle outer name. The middle part tells us that the inner name x2 is connected to the edge e3, and x3 is linked to the outer name y1.
To sum up, we rewrite edge identifiers in the order as they are revealed when traversing the place graph. The complete link recording procedure is then, to first record all idle inner names, then inner names connected to edges, after connections from inner to outer names, lastly idle outer names. Recall that idle edges are not captured as we treat only lean bigraphs (cf. Def. 9).
Summary. The basic schema of the encoding resembles the following form: \(PE_0\#\cdots \#PE_{m}\#LE\#\), where \(PE_i\) denotes the place encoding for the ith root (\(i \in m\)) of \(B^P\) such that the natural order is preserved,Footnote 3 and LE denotes the link encoding.
Examples
Unary interfaces. We provide more examples to illustrate the algorithm. The breadth-first string encodings for each of the four bigraphs in Fig. 2 are:
-
(a)


-
(b)


-
(c)


-
(d)


Imagine, we would extract the corresponding encoding of the link part, the result would be for bigraph (c) D{e0}E{e0e1}F{e1}D{e2}E{e3}F{e2e3}, for (d) D{e0}E{e0e1}F{e1}D{e2e3}E{e2}F{e3}. Note that the link part cannot be obtained without traversing the place graph since we need to respect the structural interplay imposed by both the place and link graph. Both (c) and (d) exhibit the same place graph, however, the linkings between the nodes are slightly different which is visible in the encoding above. Also bigraphs (a) and (b) have a similar structure. Until the third level of the place graph, the encoding seems the same. Note the last appearing control F which is connected to y2 in (a) and to y1 in (b). Another difference is the end of the encoding in (a) compared to (b). Here, we see that the inner name b is connected to the outer name y2 which is not the case in (b) (see Fig. 2).
Elementary bigraphs. Now, we consider elementary bigraphs, a special class of node-free bigraphs that can be classified as placings \(\phi\) and linkings \(\lambda\) [33]. Table 1 shows their canonical encoding. Elementary bigraphs represent a set of basic bigraphs of which more complex ones can be built.
We see that \({\mathrm{BFSE}}({merge}_2)\) and \({\mathrm{BFSE}}(join)\) is equal to  which makes sense because \(merge_n\) itself is recursively defined using only the identity place graph at 1 and join in the following manner: \({merge}_0 = 1, {merge}_1 = {\mathsf {id}}_1, {merge}_2 = join, merge_{n+1} = join \circ ({\mathsf {id}}_1 \otimes {merge}_n)\) (cf. [33]). Concerning the linkings \(\lambda\) in Table 1, the symbol \(\epsilon\) is called the origin, a trivial interface defined as
which makes sense because \(merge_n\) itself is recursively defined using only the identity place graph at 1 and join in the following manner: \({merge}_0 = 1, {merge}_1 = {\mathsf {id}}_1, {merge}_2 = join, merge_{n+1} = join \circ ({\mathsf {id}}_1 \otimes {merge}_n)\) (cf. [33]). Concerning the linkings \(\lambda\) in Table 1, the symbol \(\epsilon\) is called the origin, a trivial interface defined as  . A substitution \(\sigma :X \rightarrow Y\) is the tensor product of elementary substitutions
. A substitution \(\sigma :X \rightarrow Y\) is the tensor product of elementary substitutions  , where \(Y = \{\vec{y}\}\) and \(X_0 \uplus \cdots \uplus X_{n-1}\), and a closure is the tensor product of elementary closures
, where \(Y = \{\vec{y}\}\) and \(X_0 \uplus \cdots \uplus X_{n-1}\), and a closure is the tensor product of elementary closures  , where \(W = \{\vec{w}\}\) (see Table 1 and also [33, p. 29]). Generally, a bijection from sites to roots is called a permutation \(\pi\) and a bijective substitution is called renaming \(\alpha\).
, where \(W = \{\vec{w}\}\) (see Table 1 and also [33, p. 29]). Generally, a bijection from sites to roots is called a permutation \(\pi\) and a bijective substitution is called renaming \(\alpha\).
Apparently, most of their canonical forms exhibit a finalized, static nature, i.e., the time complexity is O(1), or at least follow a simple recipe on how to construct them.
Multiary interfaces. We proceed the demonstration using bigraphs with multiary outer faces (refer to Def. 4 and Fig. 3). Particularly, bigraphs that have more than one root. First, we give the string encoding of the five bigraphs depicted in Fig. 3:
-
(a)

-
(b)

-
(c)

-
(d)
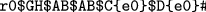
-
(e)

The only difference between bigraph \(B:\epsilon \rightarrow \langle 2, \emptyset \rangle\) (b) and \(C:\epsilon \rightarrow \langle 2, \emptyset \rangle\) (c) is that only the two trees are placed under different root indices for each bigraph. Though, bigraph B and C seem structurally similar because only the root index for each tree is different, they are not equal. Our reason to keep the original order of root nodes imposed by their indices was not arbitrary (cf. Summary in Sec. 3.1). Consider the following example, where bigraph \(A:\langle 2,\emptyset \rangle \rightarrow \langle 1, \emptyset \rangle\) (a) from Fig. 3 is used for composition, thus, interfaces as in Def. 4 are required. As per definition (cf. [33, Def. 2.5]), \(A \circ B\) is defined when the outer face of B is equal to the inner face of A, which is true in this case. The same applies to \(A \circ C\). The result of \(D = A \circ B\) is depicted in bigraph \(D:\epsilon \rightarrow \langle 1, \emptyset \rangle\) (d) in Fig. 3, and for \(E = A \circ C\) in bigraph \(E:\epsilon \rightarrow \langle 1, \emptyset \rangle\) (e). Due to \(B \,/\!\!\!{\bumpeq}\,C\) it follows \(D \,\!\!\!{\bumpeq} \, E\).
See also the discussion in “Discussion”, where we explore this issue in more detail concerning the primary usage of the proposed canonical form and other operations.
Correctness
Theorem 1
Let F and G be pure and lean bigraphs over the same signature \({\varSigma }\) with an atomic alphabet, \({\tilde{f}}\) and \({\tilde{g}}\) their corresponding bigraphical canonical string encoding. Then, \(F \Bumpeq G\) if and only if \({\tilde{f}}_i = {\tilde{g}}_i, i=\{1,\ldots ,n\}\), where n denotes the length of the string. According to Def. 10, we also say that F is isomorphic to G.
Proof outline. It is easy to verify that for two lean-support equivalent (i.e., isomorphic) bigraphs, both their BFSE must be identical. The results of the BFCF for rooted unordered trees presented in [15] will suffice to justify our proof outline, as we did not alter the fundamental algorithmic concepts. Due to the orthogonality of a bigraph’s place and link structure, we can define a string encoding of both constituents separately but not independently, assuming that both encodings are uniquely derived for some bigraph and are not necessarily a canonical form on its own. Since the “place encoding” is unique, all subtrees are in the correct forms regardless of the encoding of the link part (cf. [15]). This is also true if we include the encoding of the link part of some BFSE into our consideration. We introduce a new symbol with a smaller order among all other symbols of the alphabet with which we encode the corresponding link names connected to a node according to the lexicographic order. Then, merging both encodings is possible without contradiction, since the link part has a designated order in relation to the place part. In our approach, the link encoding is strongly connected to the place graph and depends on its ordering. Thus, if the encodings of two place graphs are not equal, both its “included” link encodings are also different with respect to their position in the string.
With regard to the bigraph isomorphism test, we say that two bigraphs A and B over the same signature are equivalent to each other, denoted by \(A \bumpeq B\), if they represent the same bigraph, or \(A \Bumpeq B\), if both bigraphs are lean. The uniqueness of the BFSE ensures a bijection in the sense that it forms a support translation, meaning, when two BFSEs \({\tilde{a}}\) and \({\tilde{b}}\) are compared character by character and for every \({\tilde{a}}_i = {\tilde{b}}_i, i \in 0,\ldots ,n\), two bigraphs are said to be equal. \(\hfill\square\)
Experimental Evaluation on the Time Complexity
This section presents the experimental results of some performance benchmarks that were conducted using our own implementation.Footnote 4 Note that this evaluation considers lean bigraphs, for which we additionally discuss sufficient reasons in "Reflections on the Application”.
To evaluate the time complexity of our algorithm, we try to follow the “common practice” of the recent bigraph-related literature [16, 23] concerning experimental evaluation. The purpose is not to compare these works with our results (which is not possible nor fair due to the different problems treated) but rather to highlight the experimental setups on the time complexity analysis. In [16] the running time of the CSP resolution and construction of the embedding of their presented approach are individually measured. The performance changes are further evaluated by adjusting the size of the bigraphs by three parameters (number of “zones”, “cars” and “connectivity degree”, cf. [16, p. 53]). In [23], four experiments are conducted concerning their bigraph matching approach. The authors measured the running time using different sizes of the redexes and agents, and by increasing the depth of the agent. The approach is compared with another library for bigraphs. Due to the fact that there are no widely recognized benchmarks for the bigraph matching or the bigraph isomorphism problem (cf. [25, 31]), the experimental setups differentiate among the works.
Now proceeding with the empirical evaluation of the implementation, we employed the test framework called Java Microbenchmark Harness (JMH) to increase the soundness. The tests described in the following were performed on a 2 Core Intel Core i7-7500U processor (3.5 GHz) with 50 warm-up iterations and 30 measurement iterations across two forks. Two different test cases were designed. In the first case, we only considered place graphs of varying size. For the second case also the link graph was included and the number of connections among the nodes were scaled. With respect to test data, we used the random bigraph generation algorithm as presented in [25]. Note that the following running times were consistently measured by JMH.
For the first test, we randomly generated place graphs with preferential attachment (see [25]), and varied the number of nodes \(n = \{1000,\ldots ,10000\}\), further fixing the root size to \(r=1\). In accordance with [15], we achieve the same complexity as the BFCF for rooted unordered trees, when only place graphs are considered. This can be observed in Fig. 4, where we normalized the measured time between 0 and 1. The reason is that the total execution time depends on the implementation (i.e., the used platform and programming language), thus, only the curve’s behavior is of interest for our analysis.
Proposition 1
The time complexity of Algorithm 2 to compute the BFSE of a place graph \(B^P\) is \(O(n^{2}\,d\log {d})\), where n is the number of places, and d the maximal node degree in \(B^P\).
Regarding the second test, we randomly synthesized bigraphs, where we also allowed connections between nodes. In [25], two strategies were proposed; here we had chosen the maximal degree variant with assortative behavior. This means that nodes with the same arity will connect to nodes with similar arity. For the test, we fixed the number of nodes to \(n=5000\) and used the signatures \({\varSigma }_i = \{{\mathsf {A}}:k_i\}\) with \(k = \{5, 10, 20\}\). More specifically, for every run \(i \in \{1,2,3\}\) exactly n nodes with the control A were created, each with \(k_i\) ports connected to \(k_i\) distinctive links. In every run i, we incrementally adjusted the fraction of the n nodes being selected for linking using \(p = \{0.1, 0.25, 0.5, 0.75, 1.0\}\). Thus, the number of possible links gradually increases.
Time complexity of the canonization algorithm for bigraphs with links. The points denote our experimental data generated for bigraphs over three different signatures with varying arity. Every bigraph contained 5000 nodes. The x-axis denotes the proportion of nodes being selected for linking. A log scale is used for the y-axis, where the time is normalized between 0 and 1
The results are presented in Fig. 5, where the measured time was normalized in the same manner as above, additionally using a logarithmic scale on the y-axis. The logarithmic scale allows us to compare the percentage changes of the running time also among the three signatures. The different colors in the legend represent the three different signatures (from \({\varSigma }_1\) to \({\varSigma }_3\)). For instance, the turquoise line represents the measurements for bigraphs over \({\varSigma }_3 = \{{\mathsf {A}}:20\}\) for all \(p_j, j=1,\ldots ,5\), e.g., at \(p_5=1.0\) every node of the bigraph had 20 ports, which were all connected to a node via a distinct link. From Fig. 5 it can be seen that for all signatures there is an increase about roughly 50% w.r.t. the running time when more than half of the nodes (\(p_3=0.5\)) are linked (from the link graph’s perspective) as opposed to \(p_1=0.1\), where only 10 % of the nodes are connected.
Regardless of the fraction of nodes being selected for linking, the maximal arity significantly determines the complexity, when comparing the data of the three different signature experiments. That means, for example, \({\varSigma }_3\) at \(p_1 = 0.1\) has a higher complexity than \({\varSigma }_1\) at \(p_5 = 1.0\). As shown in Fig 5 of this experiment, the time increases linearly with increasing node linkage for all three individual signatures. Thus, with regard to the rate of change concerning the running time in Fig. 5, we can treat the maximal arity of a signature roughly as a constant, because the rate of change is constant.
As a conclusion from the above, we can summarize the experimental results by the following claim:
Claim
The time complexity of Algorithm 2 to compute the BFSE of a pure, lean bigraph B over \({\varSigma }\) is \(O(n^{2}k\,d\log {d})\), where n is the number of places, k the maximal arity in \({\varSigma }\), \(k > 0\), and d the maximal node degree in \(B^P\).
Discussion
Now that we have defined and analyzed the canonical form of pure, lean bigraphs, we want to compare our work with related approaches, before we reflect on particular applications and consider some limitations.
Related Work
The graph isomorphism problem is well-addressed in the literature for several kinds of graphs. For the sake of shortness, we do not intend to give a complete overview of the graph isomorphism problem. Because we deal with a special graph abstraction, current approaches cannot directly be applied on bigraphs and are thus only superficially related to our work. At most, the findings may be applicable for the corresponding place or link graph only but not for both substructures at the same time (as also concluded in [6]). For a better understanding, however, we wish to present selected approaches for each domain that treat the isomorphism problem in general, before we address canonical labelings and bigraph matching implementations, and compare them to our work.
Berge and Rado [9] gave some notes on the formal construction of isomorphic hypergraphs with proofs. A quasi-polynomial time algorithm for the graph isomorphism problem of general graphs was given by Babai [2, 3]. When restricting graphs to trees, easier and less complex solutions are possible, e.g., [28, 42] both treat the subgraph isomorphism problem for rooted unordered trees. Further is [29] a serious addition in the field of graph isomorphism in polynomial time for graphs of bounded degree. Subforest isomorphism is another special case of the subgraph isomorphism problem. Given a tree G, the task is to find the forest H in G. Generally, the problem is NP-complete, however, the findings of [6] show that it is practically solvable when the number r of roots of the forest H is small (i.e., \(r \le 3\)). The exponential explosion only depends on r [6]. The graph isomorphism problem is rated as difficult in the literature. Graph isomorphism is either P or NP-complete [22]. We refer the reader to a recent work of Babai [2] for a more comprehensive elaboration on the hardness of graph isomorphism.
Canonization of graphs. In relation to our work, we use a canonical form based on the BFCF as described in [15] (refer to “Bigraphical Canonical String Encoding”). Moreover, Chi et al. emphasize that their additionally presented DFCF for labeled rooted ordered trees is equivalent to the depth-first traversal encodings proposed in [1, 34, 37] which we not review here. Many other canonization algorithms are proposed in the literature, where we wish to present some selected approaches [4, 28, 44].
Babai and Luks [4] present how to obtain a canonical form for graphs of bounded valence in polynomial time, and for general graphs in exponential time.
Valiente [44] exhibits a tree isomorphism code for rooted unordered trees. The code is a recursively defined integer sequence, each element separated by a comma symbol. Starting with the root node, each child is assigned a “subtree code”, which itself is a concatenation of subtree codes. The first integer of a code sequence associated with a node denotes the number of nodes of that subtree. Further, the integer sequences of each subtree are arranged according to ascending lexicographic order. Thus, nodes with many children are always moved to the right of the code. To obtain the code, a postorder traversal of the tree is performed. However, it does not apply to labeled trees, which place graphs are.
The canonical labeling problem in linear time for subtree isomorphism is expounded by [28] for rooted unordered trees with different types of labels. Luccio et al. [28] have convincingly presented in great detail that the running time of the processing and search algorithm depends on the alphabet and increases with more complex labels. The trees are first transformed, according to their alphabet, to ordered ones before the string encoding is computed. For searching the subtrees of the target tree, additional data structures are used. Specifically, bottom–up subtree searches are covered as a dictionary problem to find the occurrences for each pattern in the target tree. In contrast to us, the authors treat the subtree isomorphism problem.
Bigraph matching. Bigraph matching is the cornerstone to practically use bigraphs for experimentation and simulation. Research on bigraph matching with respect to bigraphical reactive systems (BRS) [33] is an instance of the sub-bigraph isomorphism problem. In bigraphs jargon, reaction rules (also named rewrite rules) define the behavior of a BRS. The search pattern of the reaction rule, called the redex, is to be found in the target bigraph, called the agent. Then, the match is replaced with the corresponding reactum of the rewrite rule.
There are few solutions concerned with the bigraph matching problem for various kinds of bigraphs; a non-exhaustive presentation is the following. For binding bigraphs (links have local scopes) by an inductive characterization of matching [10, 19, 24]; for directed bigraphs (which subsume pure bigraphs) [5]; for bigraphs with sharing (the place graph is a DAG) using a SAT-based algorithm [41]; for the pure case by [31] as a constraint satisfaction problem (CSP), and further an adapted reduction of the problem for directed bigraphs to a CSP [16].
Birkedal et al. proposed in [10] an inductive characterization of matching for binding bigraphs. The approach emphasizes on the soundness and completeness of the approach—a requirement for implementing correct matching algorithms [10]. They have shown how binding bigraphs are decomposed into elementary constituents (i.e., normal form of binding bigraphs) first, then matching sentences and rules are defined upon them to infer valid matching sentences. Using a logical programming language such as Prolog, the set of matching rules can be expressed conveniently, accordingly “operating by searching for inference trees” when applying these rules [10, p. 16].
The authors of [41] solve the matching problem for bigraphs with sharing by treating it as a computationally efficient SAT instance. The approach is implemented in BigraphER [40]—a tool programmed in the OCaml programming language using MiniSAT as underlying solver implementation. The algorithm employed for matching is a special SAT encoding of the subgraph isomorphism problem as discussed in [39]. As further stated in [41], the particular bigraph matching problem is an instance of the NP-complete subgraph isomorphism problem and thus some optimizations are applied such as the Tseitin transformation or exploiting the redex symmetries’ which can occur in case of multiple matchings in the agent. Interestingly, when executing a BRS, the tool stores the canonical form of states of the transition graph that is being synthesized to reduce the number of intermediate states [41]. Therefore, a SAT encoding to check two bigraphs on equality was presented in [39]. Two additional constraints are given which ought to be included in the initial set of constraints as described in their earlier work [41]. Hence, the Boolean matrix forms the canonical encoding. However, the encoding is designed for bigraphs with sharing, whereas we treat pure bigraphs, and provide a string labeling as opposed to a Boolean matrix.
Computing bigraph embeddings by translating it to a CSP is expounded in [31]. An embedding, as defined in [27], is a structure preserving map which is injective from a source to target bigraphFootnote 5 [31] (see also Def. 8). Regarding the construction of the linear equation system of the CSP, the authors define constraints for both bigraph constituents separately and also by some “gluing constraints” that respect the structural dependency imposed by the interplay of both substructures [31]. Overall, 34 constraint families are contained in the equation system, however, the problem is polynomially bounded with respect to the support of the bigraphs in question [31, p. 6]. The algorithm is implemented in jLibBigFootnote 6—a Java library using ChocoFootnote 7 as the underlying solver. In [16], the embedding problem for directed bigraphs is implemented in CSP. The results are experimentally validated by a simulation. The authors found “that the running time scales exponentially” [16, p. 54] on the one hand with respect to number of nodes and on the other hand with respect to the connectivity degree. To emphasize, [16, 31] consider the embedding problem, thus, the results cannot be compared easily with ours as we treat the bigraph isomorphism problem.
Summary. We wish to highlight that this work does not treat the general bigraph matching problem but the bigraph isomorphism problem. While the methods proposed are suitable for a great number of graphs, there are some special issues when working with bigraphs, such as considering the orthogonality of the two constituents in the computation (i.e., place graph’s sites and link graph’s hyperedges, inner names, and outer names). Each of the presented works is based on a unique combination of strategies and algorithmic techniques, exploiting the peculiarities of the corresponding structure in question.
Of course, one can take the naive approach and concatenate the results of the tree and hypergraph labeling, which must be accordingly adapted for both bigraph constituents. Then, the complexity would simply be the sum of \(O = O(B^P) + O(B^L)\). This, however, would not be a satisfactory conclusion. We have a more elegant solution with a lower time complexity compared to the naive approach by doing the hypergraph \(B^L\) labeling at the same time to some extent when traversing the place graph.
Reflections on the Application
Using our method, the bigraph isomorphism problem becomes O(n), where n is the length of the string encoding. However, we can improve the runtime of the computation by extending it to a parallel bottom–up BFS as discussed in [12]. They expound a direction-optimized BFS (i.e., a hybrid top–down and bottom–up step) and compare it with the top–down approach. A significant improvement is observed by the authors based on their experimental performance tests. Still, the parallel algorithm design could be adapted to conduct further tests.
The advantages of a canonical labeling over an isomorphism test is that the BFSE can be computed separately for bigraphs instead of checking two graphs pairwise at a time (cf. [38]). If the encodings are stored in a database, computational efficient searches are possible, and if needed, one can collate the strings at any time. This application is especially useful for building a bigraph database. Fast access and searches would be possible. Then, benchmarking of bigraph-related algorithms become straightforward and more consistent.
Why Lean Bigraphs?
What we deliberately left out in the background (“Graph-Theoretical Background”) was the capability of bigraphs to express dynamics through reactions but was recovered in “Related Work”. We wish to bring the focus on the topic BRS again, especially, using it to answer the question why we treated lean bigraphs (refer to Def. 9).
With reference to [33], bigraphs whose interfaces satisfy certain properties are separately termed. For example, ground bigraphs are bigraphs that do not have sites and inner names. A lean bigraph on the other hand has no idle edges defined. Both are fine properties for the rewriting in BRSs (see [33]). Idle edges may occur in \(/x \circ G\) if x is an idle name of G. Idle edges can be regarded invisible, meaning, they can be ignored (see [33, p. 29]). Whether to discard idle links or not depends on the context where bigraphs are used. For example, it makes sense to discard them in the case for redexes of bigraphical reaction rules because a “rule whose redex has an idle name leads to rather strange behavior, unlikely to be met in applications; we tend to regard such rules as unreasonable” [32, p. 37].
We wish to emphasize this fact because lean, ground, prime bigraphs are the agents in a BRS [33] that are reconfigured by reaction rules (refer to Bigraph Matching in “Related Work”) and which our method can process.
Bigraph Isomorphism
Recall, that a reaction is a labeled transition of the form  where
where  is regarded as a reaction relation, and \(a, a^\prime\) being some agents. Such a relation allows to synthesize a transition system, which can be thought of as a directed graph with nodes and edges. Nodes are called the states of the system, and edges represent the transitions between states designated by some L.
is regarded as a reaction relation, and \(a, a^\prime\) being some agents. Such a relation allows to synthesize a transition system, which can be thought of as a directed graph with nodes and edges. Nodes are called the states of the system, and edges represent the transitions between states designated by some L.
Bigraph isomorphism checks are essential for deriving such a transition system from a BRS through the application of reaction rules on bigraphs. Sevegnani and Calder [39] pointed out several applications for the bigraph isomorphism test for which our canonical labeling can be incorporated. For example, during a simulation, a transition system is derived. Here, the detection of two isomorphic bigraphs is essential to avoid creating the same states multiple times. Only the canonical form of a state is stored in the transition system which reduces the number of intermediate states. See, for example, [35], where a breadth-first exploration of the state space is used as a strategy for generating a transition system while detecting cycles.
Moreover, the test is fundamental to check BiLog [17] predicates (see [39]). The isomorphism test can be employed to determine all automorphisms of a bigraph, which is required “to count distinct occurrences in a stochastic BRS” [39, p. 63].
Operations
Now, we briefly outline another application of the proposed encoding for pure, lean bigraphs. Specifically, operations such as composition.
Let \(G:I \rightarrow J\) be a lean bigraph in \(^\backprime {\mathbf {BG}}({\mathcal {K}})\),Footnote 8 and \({\tilde{g}} = {\mathrm{BFSE}}(G)\) its string encoded representation in \({\mathbf {String}}\) computed by the function BFSE (Algorithm 2).
While being compatible to the formalization used in [23] now, consider the faithful functor named \({\mathrm{BFSE}}:{}\) \(^\backprime {\mathbf {BG}}({\mathcal {K}}) \rightarrow {\mathbf {String}}\) on the morphisms of bigraphs such that controls \(ctrl_G\), nodes and hyperedges, and both structures \(prnt_G\) and \(link_G\) are preserved (Def. 6), and further BFSE is defined on the interfaces of bigraphs. Thus, we may say that the encoding translates the bigraph G from \(^\backprime {\mathbf {BG}}({\mathcal {K}})\) into \({\mathrm{BFSE}}(G):{\mathrm{BFSE}}(I) \rightarrow {\mathrm{BFSE}}(J)\). For that purpose a functor BFSE is obtained in a manner that allows to move back and forth between the interface of some bigraph G and the corresponding index in its string encoding \({\tilde{g}}\). For example, \({\mathrm{BFSE}}(J) = j\), where j constitutes a partially ordered set of indices of length \(m + |Y|\), the first m elements of j are indices that represent the position of root symbols in the encoding, similarly, |Y| elements map to the index of outer name symbols in \({\tilde{g}}\). Considering bigraph (a) in Fig. 3, \(A:\langle 2,\emptyset \rangle \rightarrow \langle 1,\emptyset \rangle\) with  , its outer face image \({\mathrm{BFSE}}(J)\) is the set \(\{0\}\), and analogously, \({\mathrm{BFSE}}(I) = \{6,8\}\).
, its outer face image \({\mathrm{BFSE}}(J)\) is the set \(\{0\}\), and analogously, \({\mathrm{BFSE}}(I) = \{6,8\}\).
By looking at the method’s definition, identity and composition are preserved. If BFSE preserves composition, \({\mathrm{BFSE}}(G \circ F) = {\mathrm{BFSE}}(G) \circ {\mathrm{BFSE}}(F)\). However, we refrain from giving the exact details in this descriptive presentation here as it is out of scope in this paper. A proof of whether composition is preserved for all morphisms in \(^\backprime {\mathbf {BG}}({\mathcal {K}})\) is interesting future work.
Atomicity of the Alphabet
Recall that the signature of two bigraphs must be the same, so that these can be equal at all (refer to Theorem 1). Even so, the control labels must satisfy a certain condition. Our method assumes that the signature’s control labels are drawn from an atomic alphabet. Thus, it cannot be directly applied to other alphabets. Consider the following problem with a different, non-atomic alphabet. Let us assume we have the signature \({\varSigma }= \{{\mathsf {L}}:0, {\mathsf {LL}}:0\}\), a bigraph F with two nodes \(ctrl(v_1) = {\mathsf {LL}}\) and \(ctrl(v_2) = {\mathsf {L}}\) under the root, and G with three nodes \(ctrl(v_1) = {\mathsf {L}}\), \(ctrl(v_2) = {\mathsf {L}}\) and \(ctrl(v_3) = {\mathsf {L}}\) under the root. Their corresponding BFSE is in both cases  , though G has more children then F. Fig. 6 illustrates that non-atomic alphabets violate Theorem 1. Disregarding that property would result that \(F \Bumpeq G\), which is not true and can be seen without further proof in Fig. 6.
, though G has more children then F. Fig. 6 illustrates that non-atomic alphabets violate Theorem 1. Disregarding that property would result that \(F \Bumpeq G\), which is not true and can be seen without further proof in Fig. 6.
Two bigraphs F, G defined over the same signature \({\varSigma }= \{{\mathsf {L}}:0, {\mathsf {LL}}:0\}\). Non-atomic alphabets violate Theorem 1
A solution is presented in [28], where the authors explain how to treat the ordering of labeled trees with non-constant alphabets. Notice that non-constant alphabets increase the string size and thus the complexity of the preprocessing and search. For two bigraphs F and G, the running time is then a function of the total length of all control labels in both bigraphs (cf. [28]).
Another approach to treat non-atomic alphabets is by parametrization of control labels. For instance, this is accomplished by a suitable hash function that generates a unique integer for each distinguished control label. Hashes need to be computed only once and can be stored in a map. Hence, accessing the hashed value is computationally efficient and takes constant time. Concerning our method, in the course of the place graph traversal and ordering, one would then acquire the corresponding hashed value of the control label instead of using the label directly. Consequently, the comparison of two string encodings has to be slightly changed. The encoding itself is not a pure string anymore but rather an array. With the exception of the special symbols (e.g.,  or #), the computed values of the hash function would be allocated to each cell in the array corresponding to the respective control at that position. Comparing the array is then a matter of comparing the hashed values for the array’s same indices.
or #), the computed values of the hash function would be allocated to each cell in the array corresponding to the respective control at that position. Comparing the array is then a matter of comparing the hashed values for the array’s same indices.
Special attention must be paid to ensure that the hashing operation is computationally inexpensive and takes account of collision and dispersion qualities. Given the fact that non-atomic alphabets may increase the running time of the encoding in some circumstances, their use shall be carefully considered. So far we did not have encountered a problem with atomic alphabets in practice. However, it would be interesting future work to consider alternative approaches in which the encoding could handle non-atomic alphabets.
Conclusion
During the canonical encoding procedure, each bigraph is assigned a unique string based on its place and link graph. To determine whether two bigraphs are equal, i.e., isomorphic to each other, or (lean-) support equivalent (see Defs. 8, 9 and 10), one only has to compare the strings with each other instead of traversing the bigraphs completely.
As a result, we casted the bigraph isomorphism problem to a string matching problem by testing two bigraphical canonical forms, whether they are equal. This was achieved by defining a unique string encoding, which reduces a bigraph’s place graph to the case of an ordered tree. The proposed solution was devised for pure and lean bigraphs, specifically, for bigraphs with unary or multiary interfaces, and no idle edges. The encoding guarantees lean-support equivalence (see Def. 9), which means that our canonical mapping is a structure-preserving mapping, if two lean bigraphs are equal in the sense that their string encodings are equal. Our approach assumes atomic alphabets. It seems that non-atomic alphabets are possible in general, however, they were not needed for the approach presented in this work and is thus left for future work.
Notice that even if the orthogonality of the bigraph allows separate handling of both substructures, we presented a solution that obtains the canonical form by traversing the place graph and the link graph at the same time to some extent. We showed that the complexity of our canonization algorithm is not significantly higher than the one of the BFCF for rooted unordered trees in [15]. The complexity of our method for a bigraph B over \({\varSigma }\) is \(O(n^2\,k\,d\log {d})\), where n is the number of nodes, k the maximal arity of a bigraph’s signature and d the maximal node degree of the place graph. This claim is supported by experiments.
Availability of data and material
Not applicable.
Code availability
Code is available currently upon request through the corresponding author. More details can be obtained in the future from https://www.bigraphs.org/.
Notes
The concept of interfaces for graphs is not new and are also prominent in gs-graphs [11] and ranked graphs [21], as they conveniently allow the definition of graph composition [11]. This is, however, not within the scope of this paper. For a detailed explanation of bigraph composition, the reader may refer to [33].
When we refer to idle inner names, we actually mean closures, as introduced in “Examples” later.
The authors are working on a framework for the creation and simulation of bigraphs, called Bigraph Framework which is implemented in the Java programming language. The proposed algorithm is implemented in this framework. More details can be obtained from https://www.bigraphs.org/.
Specifically, this can be regarded as a “weak” support translation “from nodes and edges of the redex to nodes and edges of the agent” [31, p. 2].
For more information, we refer to this URL: http://midas.uniud.it/downloads/libbig/.
Choco [36] is a open-source Java library for constraint programming.
Categorically, bigraphs and their interfaces are classified in the \(^\backprime {\mathbf {BG}}({\mathcal {K}})\) category which has arrows that are bigraphs and objects that are bigraph interfaces [33].
References
Asai T, Arimura H, Uno T, Si Nakano. Discovering frequent substructures in large unordered trees. In: Grieser G, Tanaka Y, Yamamoto A, editors. Discovery science, lecture notes in computer science. Berlin: Springer; 2003. p. 47–61. https://doi.org/10.1007/978-3-540-39644-4_6.
Babai L. Graph isomorphism in quasipolynomial time. CoRR. 2015. arxiv:1512.03547.
Babai L. Graph isomorphism in quasipolynomial time [Extended Abstract]. In: Proceedings of the Forty-Eighth Annual ACM Symposium on Theory of Computing, STOC ’16. ACM; 2016. p. 684–97. https://doi.org/10.1145/2897518.2897542.
Babai L, Luks EM. Canonical labeling of graphs. In: Proceedings of the Fifteenth Annual ACM Symposium on Theory of Computing, STOC ’83. ACM; 1983. p. 171–83. https://doi.org/10.1145/800061.808746.
Bacci G, Grohmann D, Miculan M. DBtk: a toolkit for directed bigraphs. In: Proceedings of the 3rd International Conference on Algebra and Coalgebra in Computer Science, CALCO’09. Springer-Verlag; 2009. p. 413–22. http://dl.acm.org/citation.cfm?id=1812941.1812978.
Bacci G, Miculan M, Rizzi R. Finding a forest in a tree. In: Maffei M, Tuosto E, editors. Trustworthy global computing, lecture notes in computer science. Berlin: Springer; 2014. p. 17–33.
Beamer S, Asanović K, Patterson D. Direction-optimizing breadth-first search. In: Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, SC ’12. IEEE Computer Society Press; 2012. p. 12:1–12:10.
Beamer S, Asanović K, Patterson DA. Searching for a parent instead of fighting over children: a fast breadth-first search implementation for Graph500. 2011. Technical Report, UCB/EECS-2011-117, EECS Department, University of California, Berkeley. URL http://www2.eecs.berkeley.edu/Pubs/TechRpts/2011/EECS-2011-117.html.
Berge C, Rado R. Note on isomorphic hypergraphs and some extensions of Whitney’s theorem to families of sets. J Combin Theory Ser B. 1972;13(3):226–41. https://doi.org/10.1016/0095-8956(72)90058-5.
Birkedal L, Damgaard TC, Glenstrup AJ, Milner R. Matching of bigraphs. Electron Notes Theor Comput Sci. 2007;175(4):3–19. https://doi.org/10.1016/j.entcs.2007.04.013.
Bruni R, Montanari U, Plotkin G, Terreni D. On hierarchical graphs: reconciling bigraphs, Gs-monoidal theories and Gs-graphs. Fundamenta Informaticae. 2014;134:287–317. https://doi.org/10.3233/FI-2014-1103.
Buluc A, Beamer S, Madduri K, Asanovic K, Patterson D. Distributed-memory breadth-first search on massive graphs. CoRR. 2017. arxiv:1705.04590.
Bunke H, Dickinson P, Kraetzl M. Theoretical and algorithmic framework for hypergraph matching. In: Roli F, Vitulano S, editors. Image analysis and processing–ICIAP 2005, vol. 3617. Berlin: Springer; 2005. p. 463–70. https://doi.org/10.1007/11553595_57.
Chi Y, Muntz RR, Nijssen S, Kok JN. Frequent subtree mining—an overview. Fundamenta Informaticae. 2005;66(1–2):161–98.
Chi Y, Yang Y, Muntz RR. Canonical forms for labelled trees and their applications in frequent subtree mining. Knowl Inf Syst. 2005;8(2):203–34. https://doi.org/10.1007/s10115-004-0180-7.
Chiapperini A, Miculan M, Peressotti M. Computing embeddings of directed bigraphs. In: Gadducci F, Kehrer T, editors. Graph transformation, lecture notes in computer science. Springer International Publishing: Berlin; 2020. p. 38–56. https://doi.org/10.1007/978-3-030-51372-6_3.
Conforti G, Macedonio D, Sassone V. Spatial logics for bigraphs. In: Caires L, Italiano GF, Monteiro L, Palamidessi C, Yung M, editors. Automata, languages and programming, lecture notes in computer science. Berlin: Springer; 2005. p. 766–78.
Conte D, Foggia P, Sansone C, Vento M. Thirty years of graph matching in pattern recognition. Int J Pattern Recognit Artif Intell. 2004;18(03):265–98. https://doi.org/10.1142/S0218001404003228.
Damgaard TC, Glenstrup AJ, Birkedal L, Milner R. An inductive characterization of matching in binding bigraphs. Formal Aspects Comput. 2013;25(2):257–88. https://doi.org/10.1007/s00165-011-0184-5.
Ehrig H, Ehrig K, Prange U, Taentzer G. Fundamentals of algebraic graph transformation. In: Monographs in theoretical computer science. An EATCS series. Berlin: Springer; 2006.
Gadducci F, Heckel R. An inductive view of graph transformation. In: Presicce FP, editor. Recent trends in algebraic development techniques, lecture notes in computer science. Berlin: Springer; 1998. p. 223–37. https://doi.org/10.1007/3-540-64299-4_36.
Garey MR, Johnson DS. Computers and intractability: a guide to the theory of np-completeness. New York: W. H. Freeman & Co; 1979.
Gassara A, Bouassida Rodriguez I, Jmaiel M, Drira K. Executing bigraphical reactive systems. Discrete Appl Math. 2019;253:73–92. https://doi.org/10.1016/j.dam.2018.07.006.
Glenstrup AJ, Damgaard TC, Birkedal L, Højsgaard E. An implementation of bigraph matching. 2010. Technical Report, TR-2010-135, IT University of Copenhagen, Denmark. ISSN 1600–6100. ISBN 978–87–7949–228–8.
Grzelak D, Priwitzer B, Aßmann U. Generating random bigraphs with preferential attachment. CoRR. 2020. arxiv:2002.07448.
Hartke S, Radcliffe A. McKay’s canonical graph labeling algorithm. Contemp Math Book Ser. 2013. https://doi.org/10.1090/conm/479/09345.
Højsgaard E. Bigraphical languages and their simulation. 2012. PhD Dissertation. https://core.ac.uk/download/pdf/50526631.pdf.
Luccio F, Mesa Enriquez A, Olivares Rieumont P, Pagli L. Bottom-up subtree isomorphism for unordered labeled trees. 2004. Technical Report, TR-04-13, Dipartimento di Informatica, Università di Pisa, Italy. http://compass2.di.unipi.it/TR/files/TR-04-13.ps.gz.
Luks EM. Isomorphism of graphs of bounded valence can be tested in polynomial time. In: 21st Annual Symposium on Foundations of Computer Science (Sfcs 1980). 1980. p. 42–9. https://doi.org/10.1109/SFCS.1980.24.
McKay BD, Piperno A. Practical graph isomorphism, II. J Symb Computat. 2014;60:94–112. https://doi.org/10.1016/j.jsc.2013.09.003.
Miculan M, Peressotti M. A CSP implementation of the bigraph embedding problem. CoRR. 2014. arxiv:1412.1042.
Milner R. Bigraphical reactive systems: basic theory. 2001. Technical Report UCAM-CL-TR-523, University of Cambridge, Computer Laboratory. ISSN 1476-2986.
Milner R. The space and motion of communicating agents. 1st ed. Cambridge: Cambridge University Press; 2009.
Nijssen S, Kok JN. Efficient discovery of frequent unordered trees. In: In First International Workshop on Mining Graphs, Trees and Sequences. 2003. p. 55–64.
Perrone G. Domain-specific modelling languages in bigraphs; 2013. PhD Dissertation, IT University of Copenhagen, Denmark.
Prud’homme C, Fages JG, Lorca X. Choco solver documentation. TASC, INRIA Rennes, LINA CNRS UMR 6241, COSLING S.A.S. 2016. http://www.choco-solver.org.
Rückert U, Kramer S. Frequent free tree discovery in graph data. In: Proceedings of the 2004 ACM Symposium on Applied Computing, SAC ’04. ACM; 2004. p. 564–70. https://doi.org/10.1145/967900.968018.
Schweitzer P, Wiebking D. A unifying method for the design of algorithms canonizing combinatorial objects. CoRR. 2019. arxiv:1806.07466.
Sevegnani M, Calder M. Bigraphs with sharing. Theoret Comput Sci. 2015;577:43–73. https://doi.org/10.1016/j.tcs.2015.02.011.
Sevegnani M, Calder M. BigraphER: rewriting and analysis engine for bigraphs. In: Chaudhuri S, Farzan A, editors. 28th International Conference on Computer Aided Verification, vol. 9780. Springer International Publishing; 2016. p. 494–501. https://doi.org/10.1007/978-3-319-41540-6_27.
Sevegnani M, Unsworth C, Calder M. A SAT based algorithm for the matching problem in bigraphs with sharing. 2010. Technical Report. University of Glasgow. http://dcs.gla.ac.uk/~michele/papers/tech_match.pdf.
Shamir R, Tsur D. Faster subtree isomorphism. J Algorithms. 1999;33(2):267–80. https://doi.org/10.1006/jagm.1999.1044.
Ullmann JR. An algorithm for subgraph isomorphism. J ACM. 1976;23(1):31–42. https://doi.org/10.1145/321921.321925.
Valiente G. Algorithms on trees and graphs. Berlin: Springer; 2002. https://doi.org/10.1007/978-3-662-04921-1.
Wachter M, Haenni R. Propositional DAGs: a new graph-based language for representing boolean functions. In: Proceedings of the Tenth International Conference on Principles of Knowledge Representation and Reasoning, KR’06. AAAI Press; 2006. p. 277–85.
Acknowledgements
Funded by the German Research Foundation (DFG, Deutsche Forschungsgemeinschaft) as part of Germany’s Excellence Strategy—EXC 2050/1—Project ID 390696704—Cluster of Excellence “Centre for Tactile Internet with Human-in-the-Loop” (CeTI) of Technische Universität Dresden.
Funding
Open Access funding enabled and organized by Projekt DEAL. Funded by the German Research Foundation (DFG, Deutsche Forschungsgemeinschaft) as part of Germany’s Excellence Strategy—EXC 2050/1—Project ID 390696704—Cluster of Excellence “Centre for Tactile Internet with Human-in-the-Loop” (CeTI) of Technische Universität Dresden.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
D. Grzelak and U. Aßmann are also with the Centre for Tactile Internet with Human-in-the-Loop (CeTI), Technische Universität Dresden, 01062 Dresden, Germany.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Grzelak, D., Aßmann, U. A Canonical String Encoding for Pure Bigraphs. SN COMPUT. SCI. 2, 246 (2021). https://doi.org/10.1007/s42979-021-00552-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-021-00552-5





 is the set of ports of F.
is the set of ports of F.