Abstract
The aim of this study was to analyse the level and structure of genetic and morphometric variation in the East-Central European populations of Phengaris (Maculinea) nausithous. On the basis of the life cycle of the species, we expected a low level of variation within the populations coupled with a relatively high level of differentiation among them. We also assumed that the differentiation among the populations has a regional pattern. In order to confirm these assumptions, we collected population samples from two regions within the Carpathian Basin (western Transdanubia and Transylvania) and from a region east of the Carpathian Mountains (Bukovina). The level of enzyme polymorphism and the amount of morphometric variation were investigated in the sampled populations. As the western (Transdanubia) and the eastern populations (Transylvania plus Bukovina) were suggested to belong to different subspecies (Rákosy et al. in Nota Lepidopterol 33:31–37, 2010), our working hypothesis was that these two groups of populations belong to different evolutionary significant units (ESUs). The results of all genetic analyses (PCA, Bayesian-clustering analysis and especially the UPGMA dendrogram) confirmed our assumption. The level of differentiation was higher between the western and eastern populations resulting in two monophyletic lineages of Phengaris (Maculinea) nausithous in East-Central Europe. This result suggests that these lineages can be considered as two different ESUs.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Professor Zoltán Varga is one of the most enthusiastic conservation biologists in Hungary. He recognised fairly early the significance of studying genetic variation in natural populations. In the late 70’s, he established a molecular population genetic laboratory in his department. At that time the study of enzyme polymorphism was the most popular molecular method all over the scientific world. The first author of this paper has been working in this laboratory from the beginning. Later, generations of PhD students were educated there. The main goal of Z. Varga, on the one hand, was to carry out various research projects in natural populations of endangered or vulnerable butterfly species; but, on the other hand, he intended to provide data for everyday nature conservation on the level and structure of genetic variation in these species. His aims were completely fulfilled and the results obtained in this population genetic laboratory were published in several scientific papers. The present study was also carried out in this laboratory. Moreover, some aspects of the results, especially regarding the Phengaris (Maculinea) species were used in practical nature conservation and helped to work out conservation strategies for these species.
In order to develop efficient conservation strategies for endangered butterfly species, it is of great importance to outline proper conservation units (Allendorf and Luikart 2009). The two most commonly discussed conservation units are evolutionarily significant unit (ESU) and management unit (MU) or functional conservation unit (FCU) (Funk et al. 2012). Out of these, the ESU was introduced by Ryder (1986) and characterised as a group of genetically and ecologically differentiated populations. Nonetheless, the definition of an ESU has changed over the past three decades. Moritz (1994) regarded this as a reciprocally monophyletic unit. A further operational concept of an ESU was introduced by Crandall et al. (2000) who emphasised the importance of ecological and genetic differentiation between different entities. With this in mind, the other important unit in conservation biology is FCU or MU, a group of demographically and ecologically independent populations (Maes et al. 2004; Allendorf and Luikart 2009; Funk et al. 2012). Thus, FCUs/MUs are often defined within ESUs and represent populations that are important for the long-term persistence of an ESU (Hughes et al. 1997; Vila et al. 2006; Funk et al. 2012). This implies that FCUs must be genetically, and perhaps adaptively, differentiated within an ESU (Palsbøll et al. 2007; Funk et al. 2012); it is therefore of utmost importance in practical nature conservation to delineate the FCUs of endangered species.
The dusky large blue Phengaris (Maculinea) nausithous (Bergsträsser 1779) has a highly specialised life cycle. Imagoes lay eggs on the larger, apical flower heads of Sanguisorba officinalis (Thomas 1984). Larvae develop through three instars in the flowers and green fruits (Elmes et al. 1991; Thomas and Wardlaw 1992). Then they drop on the ground and wait for being discovered by the foraging workers of the ant Myrmica rubra (Thomas 1984; Thomas et al. 1989; Tartally et al. 2008) who adopt and transport them to the ant nest. Caterpillars live there as social parasites for about 10 month during the fourth instar (Elmes et al. 1991). The species has a Euro-Siberian distribution with a wide but sporadic range from Western Europe through Southern Siberia to Mongolia (Lukhtanov and Lukhtanov 1994; Tuzov 1997; Wynhoff 1998; Munguira and Martin 1999; Rákosy et al 2010). The European distribution of the species has a definite gap in the Carpathian Basin. It is locally frequent in the western and northern hilly regions of Transdanubia, but was known to be completely absent in the Pannonian lowland as well as in Transylvania (Rákosy et al. 2010). However, P. nausithous has recently been discovered in Transylvania near Cluj-Napoca at Răscruci and Fânaţele Clujului (Rákosy et al. 2010). The ecology and host ant use of these two isolated populations are different from the Central European ones. Moreover, their appearance coincides with the original description of P. nausithous kijevensis (Sheljuzhko 1928). Therefore, Rákosy et al. (2010) suggested that the two Transylvanian populations belong to P. nausithous kijevensis.
Since P. nausithous is dependent on the presence of two sequential resources and lives in strictly localised small populations (Thomas 1984; Thomas et al. 1989), habitat fragmentation and isolation resulted in severe decline in many of its populations. Therefore, the aim of our study was to analyse the level and pattern of genetic and phenotypic differentiation among the P. nausithous populations of the Carpathian Basin and those of Bukovina, east of the Carpathian Mountains. We were especially interested in the differentiation between the populations located west (western Transdanubia) and east (Transylvania and Bukovina) of the Pannonian lowland, i.e. of the gap in the distribution of the species. Based on the pattern of differentiation among these populations, we intended to identify the conservation units of P. nausithous in East-Central Europe. Our working hypothesis was that the eastern populations belong to a different subspecies (Rákosy et al. 2010), therefore these populations can be considered as different ESU-s. In agreement with our aims, we collected population samples from a western region (Western Transdanubia) and two eastern regions (Transylvania and Bukovina) and analysed the level enzyme polymorphism together with the amount of morphometric variation in these samples.
Materials and methods
Samples
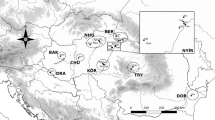
P. nausithous samples originated from 3 regions: Hungary (Őrség region in Western Transdanubia: NYDT), Transylvania (close to Cluj Napoca: TRY) and Bukovina (east of the Carpathian Mountains: BUK). In the Transdanubian region we had 6, in Transylvania 3, and in Bukovina also 3 population samples (Table 1 and Fig. 1). On average, we obtained about 24 individuals per sample though the sample sizes varied greatly. In order not to damage the sampled populations mostly males were collected at the end of the flight period after the females laid their eggs. After collection, the individuals were immediately frozen and kept at − 80 centigrade until electrophoresis.
M. nausithous samples in the Carpathian Basin and in Bukovina. Western Transdanubia (NYDT), Nagygörbő (Ng), Orfalu (Orf), Szomoróc (Szo), Őriszentpéter (Oszp), Magyarszombatfa (Mfa), Kétvölgy (Kv). Transylvania (TRY), Fanatele A (FanA), Fanatele B (FanB), Rascruci (Ras). Bukovina (BUK), Radauti (Rad), Solca (Sol), Sucevita (Suc)
Enzyme studies
Allozyme polymorphism was studied in all 12 populations at 17 different loci by vertical polyacrylamide gel electrophoresis: aconitase (Acon), acid phosphatase (AcphB), aldehyde oxidase (Aox), esterase (Est), glucose-6-phosphate dehydrogenase (G6pdh), glutamate dehydrogenase (Gdh), glutamate oxalacetate transaminase (GotA and GotB), α-glycerophosphate dehydrogenase (αGpdh), hexokinase (Hk), isocitrate dehydrogenase (IdhA and IdhB), malate dehydrogenase (Mdh), malic enzyme (Me), phosphoglucose isomerase (Pgi), phosphoglucomutase (Pgm) and superoxide dismutase (Sod). Thoraxes and abdomens were homogenised separately in 350–400 µl extraction buffers. Thorax samples were used to study G6pdh, GotA, GotB, αGpdh, Hk, IdhA, IdhB, Mdh, Me, Pgi, Pgm, and Sod, while abdomen extracts were used to analyse Acon, AcphB, Aox, Est and Gdh. The extraction buffer, the electrophoresis buffer systems and running conditions together with the staining solutions were slightly modified after Bereczki et al. (2005).
Morphometric studies
Before electrophoresis, the wings of the individuals were cut and analysed later separately. Not all samples were used in the morphometric analyses. Measurements were conducted for two samples from Transdanubia, for two samples from Transylvania and for one sample from Bukovina (Table 1). Wings were fixed on transparency films and photographed with a Sony DSC-H2 digital camera. Measurements were completed on high-resolution digital photographs by computer using the image J 1.36 programme (Kizic and Borovac 2001). Four traits were measured both on the forewings and the hindwings. Two traits were distance metrics describing the size of the wings, while the other two were angle metrics indicating the shape of them. Distances were determined between fix points. Anal length: between the basal end of the discal cell and the outer end of the anal veins (1A + 2A on the forewing and 3A on the hindwing); length of the outer margin: between the outer end of the radial vein (R3 on the forewing and Sc + R1 on the hindwing) and the outer end of the anal veins (1A + 2A on the forewing and 3A on the hindwing). Two angles were also measured both on the forewings and hindwings. Basal angle enclosed by the costal and anal margins and apical angle enclosed by the costal and outer margins. The morphometric study was only carried out on males, thus the sample sizes were slightly lower in these analyses than in the enzyme studies.
Statistical analyses
Statistical procedures aimed to compare the results obtained for morphological and genetic data could partly be compared. Variation was studied in two ways: both the amount and the structure of it were analysed.
Genetic data
Genotype and allele frequencies were calculated on the basis of banding patterns. Measures of genetic variation were calculated for each population sample using genalex 6.5 (Peakall and Smouse 2006) and fstat v.1.2 (Goudet 1995): average number of alleles per locus (nA), allelic richness (Ar), proportion of fixed alleles (%Fix), average observed heterozygosity (Ho) and proportion of polymorphic loci using the 95% criterion (P95). These parameters were compared among the geographic regions by performing ANOVA running glim 4 (Francis et al. 1994). The genetic structure of the populations was first analysed by applying principal component analysis (PCA) using the allele frequency data of the samples. The computation of allele frequencies was performed by GenAlEx6 (Peakall and Smouse 2006), while past ver.1.56 (Hammer et al. 2001) was used to carry out PCA. As a next step, Bayesian-clustering method was used (Pritchard et al. 2000) to estimate the most probable number of genetically differentiated groups (K) in our samples and assigned the individuals to these groups. structure 2.3.2 (Pritchard et al. 2010) was run to carry out these analyses without population priors, using admixture model, an initial burn-in value of 100,000 and running length of 500,000 iterations. K was assumed between 1 and 12 repeating each run ten times. In the next step, we calculated the change in log-scaled probability between successive K values (ΔK) (Evanno et al. 2005), using structure harvester 0.6.91 (Earl and VonHoldt 2011). The distribution of total genetic variation at various levels of the hierarchy was also studied by AMOVA (Excoffier et al. 1992; Weir 1996). In this analysis, the total genetic variation was partitioned into three components: between regions (BW), among populations within a region (WR/BP), and among individuals within a population (WP). AMOVA was carried out by arlequin version 3.11 (Schneider et al. 2000). Finally an UPGMA dendrogram was constructed (Sneath and Sokal 1973) on the basis of the Cavalli-Sforza and Edwards chord distances derived from the allele frequencies of the samples (Cavalli-Sforza and Edwards 1967). The statistical support of the branching pattern in the dendrogram was estimated with 3000 nonparametric bootstrap replicates (Felsenstein 1985). The percentages of replicates where the nodes are still supported are shown by the dendrogram. past ver.1.56 (Hammer et al. 2001) was used to process the dendrogram and carry out the bootstrap.
Morphological data
Phenotypic variation of the samples was characterised by the coefficients of variation calculated for each population (Sokal and Rohlf 1981). The average values were used to compare the level of variation among the regions. Pearson product moment correlation was computed between the genetic (%Fix and nA) and morphometric (CV) measures of variation (Sokal and Rohlf 1981) using the programme glim4 (Francis et al. 1994). Multiple discriminant analysis (MDA) was computed in order to determine the most probable number of groups and to compare the distribution of canonical variates among these groups. Spss 16.0 programme package was used for the computation of this analysis. The distribution of phenotypic variation at different levels of the hierarchy was analysed by a hierarchical ANOVA using glim 4 (Francis et al. 1994). In this analysis, all traits were analysed separately and then the percentages were averaged over the traits. The levels of hierarchy were similar to those in AMOVA.
Results
Level of variation in the populations
Genetic variation characterised by different parameters of enzyme polymorphism indicated a relatively low level of variation in the P. nausithous populations (Table 2: Total). Comparing these parameters among the regions the highest values were found for the Transylvanian populations. Nevertheless, none of the regional differences proved to be significant. (Table 2: ANOVA and P).
Morphometric variation was characterised by the coefficients of variation (CV) which were averaged separately over the traits characterising the size (distances) and the shape (angles) of the wings. The average CV values computed for the size measurements were 65% higher than those processed for the angles (Fig. 2A). Moreover, the CV values of the size measurements averaged for the three regions differed considerably, indicating a lower variation of wing size in the Transylvanian region, compared to the other two ones. Nonetheless, the average CV values of the angles did not differ among the regions (Fig. 2A). These latter CV values, however, were different between the two wings, namely the averages of the hindwings were higher than those of the forewings. It thus appears that the shape of the hindwings is more variable than that of the forewings.
Average values of variation coefficients (CV) estimated for the M. nausithous populations. A CV values were averaged over the regions for the measurements indicating the size of the wings (Size), for all angles (Ang.tot.), for the angles of the forewing (Ang.fw.) and for those of the hindwing (Ang.hw.). NYDT Western Transdanubia, TRY Transylvania, BUK: Bukovina. B Correlation between the observed number of alleles (nA) and the CV values averaged over the size measurements of the wings in the five populations (R = 0.692; P = 0.196). C Correlation between the proportion of fixed alleles (%Fix) and the CV values averaged over the size measurements of the wings in the five populations (R = 0.594; P = 0.291)
We have analysed the correlation between the parameters of enzyme polymorphism and the CV values averaged over the size and the shape measurements separately. None of these analyses resulted in a significant correlation, but all associations between the measurements of enzyme polymorphism and the average CV values calculated for the size of the wings indicated a trend. Namely, as the level of genetic variation increased the amount of morphometric variation decreased. This trend was the strongest between the CV values computed for the size of the wings and the proportion of fixed alleles (Fig. 2C: %fix) as well as the average number of alleles per locus (Fig. 2B: nA).
Structure of variation
In the analysis of the genetic structure of variation, a principal component of analysis was first computed. The results showed a clear differentiation among the three regions: the populations of western Transdanubia (NYDT) were clearly separated from those of Bukovina (BUK) and Transylvania (TRY) along the first axis (Fig. 3). This axis explained 66.8% of the total variation in allele frequencies and the most significant loci contributing to it were Aox and Pgm. The populations of Transylvania and Bukovina were differentiated along the second axis which explained 19.5% of the variation and was determined by Idh and Pgi.
Next, we carried out a Bayesian-clustering analysis in order to (i) estimate the most probable number of genetic clusters (K) and (ii) to assess the importance of the geographic origin of the populations. Accordingly, we ran Structure 2.3.2 assuming K between 1 and 12 repeating each run ten times. The most probable K values were selected using Structure Harvester 0.6.91. On the basis of the ΔK values, K = 3 was estimated as the most likely number of genetic clusters in the P. nausithous populations. As a next step, the distribution of these genetic clusters was calculated for all individuals in all populations. We observed a considerable heterogeneity in the frequencies of the clusters. The distribution of the three clusters specified three genetic groups (‘genetic regions’) corresponding to the three geographic regions in the investigated populations (Fig. 4). The triangle plot of the cluster probabilities indicated a stronger differentiation between the Transdanubian populations and those of Transylvania together with Bukovina supporting the results of PCA. Moreover, certain alleles of the Aox and Pgm loci proved to be cluster specific: Pgi2 for cluster 1 (characteristic for the Transdanubian populations), while Aox4 and Aox5 for cluster 3 (typical for the populations of Bukovina).
The results of the Bayesian-clustering analyses in the M. nausithous populations. The most probable number of clusters was K = 3. NYDT Western Transdanubia, TRY Transylvania, BUK Bukovina. Cl1, Cl2 and Cl3: the assumed clusters. The columns (Upper part of the figure) and the dots (lower part of the figure) represent the cluster probabilities estimated for the individuals
Phenotypic differentiation was first analysed by a multiple discriminant analysis using the morphometric data of the individuals. The results revealed that the regions were significantly differentiated along the first root (χ2 = 49.7; DF = 20; P < 0,001). The apical angle of the forewing contributed most to this root. The distribution of the canonical variates along this root clearly showed the differentiation of the Transdanubian individuals from those of the other two regions (Fig. 5).
The next step in the analysis of genetic variation was to compute an AMOVA. The results showed that the highest portion of variation (75.1%) could be attributed to the within-sample component, defined as the variation among the individuals (Fig. 6: Gen.). A fairly high percentage of the genetic variation (23.1%) was explained by the differences among the regions indicating a clear differentiation among them (Fig. 6: Gen.).
Simultaneously, a hierarchical ANOVA was carried out on the morphometric data. In agreement with the distribution of genetic variation, these results also suggested that the differences among the individuals contributed most (90.7%) to the phenotypic variation, whereas the differences among the three regions only explained 5.6% of the total phenotypic variation (Fig. 6: Morph.). Thus, in accordance with the results of the multiple discriminant analysis, hierarchical ANOVA also indicated a clear but rather low level of morphometric differentiation among the regions.
Finally, an UPGMA dendrogram was constructed on the basis of the Cavalli-Sforza and Edwards chord distance matrix. In this analysis, three samples of Phengaris (Maculinea) teleius were used as outgroups. These outgroup samples were clustered in a separate branch of the dendrogram supported by a high bootstrap value (Fig. 7). The P. nausithous samples were clustered in two main branches of the dendrogram also supported by a high bootstrap value (Fig. 7). These two P. nausithous branches corresponded to the western and eastern geographic regions: western Transdanubia (NYDT) and Transylvania (TRY) together with Bukovina (BUK). The populations of these two eastern regions were also separated in two branches but this separation was supported by a lower bootstrap value (Fig. 7). This result, similarly to that of PCA and the Bayesian-clustering analysis, indicated a stronger differentiation between the populations situated on the West and East side of the gap in the East-Central European distribution of this species.
UPGMA dendrogram constructed on the basis of the Cavalli-Sforsa and Edwards chord distances computed for the M. nausithous populations. Three M. teleius populations were used as outgroups (black letters). NYDT Western Transdanubia (light green letters), TRY Transylvania (blue letters), BUK Bukovina (red letters). Values at the nodes of the branches indicate bootstrap percentages from 3000 iterations
Discussion
Phengaris (Maculinea) nausithous is an obligate myrmecophilous butterfly i.e. its larvae can only develop in the nests of Myrmica species (Thomas 1984; Thomas et al. 1989). In Europe, P. nausithous mostly uses M. rubra as a host ant (e.g. in Transdaubia and Bukovina) but in Transylvania it parasitizes M. scabrinodis (Tartally et al. 2008; 2019). Since females lay eggs on the food plants regardless of the distance to the nests of the host ants (Thomas and Elmes 2001; Árnyas et al. 2006), larvae drop to the ground at random. Considering that a large portion (approx. 90%) of larvae die not discovered by the ant workers we can assume that P. nausithous populations are strongly affected by genetic drift. As a consequence, a relatively low amount of variation together with a fairly high level of differentiation is expected in its populations (Pecsenye et al. 2007). Our results fulfilled these expectations as we found a relatively low level of enzyme polymorphism in the investigated populations with an average number of alleles of nA = 1.79 (average of other lycaenid species nA = 2.23 (Pecsenye et al. 2015)), and an average heterozygote frequency of Ho = 0.116 (average of other lycaenid species Ho = 0.209 (Pecsenye et al. 2015)). The low level of enzyme polymorphism was coupled with a high level of differentiation. The overall fixation index was FST = 0.195** which is close to the value estimated by Figurny-Puchalska et al. (2000) for the Polish populations (FST = 0.153**).
The results of all analyses indicated a regional pattern of differentiation in the investigated P. nausithous populations (Hollós et al. 2012). At the same time, the outcome of the PCA and the Bayesian-clustering analysis of the genetic data together with the results of the multiple discriminant analysis of the morphometric data suggested a stronger differentiation between the western (Western Transdanubia) and eastern (Transylvania plus Bukovina) regions. Based on the differences in morphology and ecology observed in the Transylvanian P. nausithous populations, Rákosy et al. (2010) suggested that these populations belong to a different subspecies, namely to P. nausithous kijevensis described by Sheljuzhko (1928). The dendrogram constructed on the basis of the Cavalli-Sforza and Edwards chord distances showed most explicitly the differentiation between the western and eastern P. nausithous regions of East-Central Europe, as a few samples of Phengaris (Maculinea) teleius, another predator Phengaris species, were also included in this analysis. Three main branches of this dendrogram were supported by a high bootstrap value: P. teleius samples, the P. nausithous populations of western Transdanubia and those of the two eastern regions (Transylvania and Bukovina). It thus appears that the western and eastern populations of P. nausithous can be described as mutually monophyletic lineages. Keeping in mind that there is a great gap in the distribution of P. nausithous in East-Central Europe which is situated in the Pannonian lowland, our results suggest that these two monophyletic lineages are not only strongly differentiated from each other but they also have distinct distribution areas. As a consequence, the western and eastern populations of this species can be considered as different Evolutionary Significant Units (ESUs) (Moritz 1994; Crandall et al. 2000) and they probably belong to different subspecies as it was proposed by Rákosy et al. (2010). These conclusions were also supported by Ritter et al. (2013) who described a high level of differentiation between the Central- and East-European P. nausithous populations on the basis of mtDNA COI sequence and microsatellite data. They also found that the genetic composition of the Transylvanian populations is similar to that of the East-European and Asian populations. Nevertheless, they also detected strong differentiation between two genetic lineages in the West-European populations of P. nausithous. The genetic differentiation of the P. nausithous populations in the Carpathian Basin also showed a geographic pattern in the study by Bereczki et al. (2015). They concluded that the mtDNA sequence data together with the results of the microsatellite and allozyme studies reflect the biogeographical history of the species, since the differentiation of the large geographic regions coincides the disjunct distribution of P. nausithous.
Furthermore, our results (especially those of the PCA and the dendrogram) also indicate a clear but a lower level of differentiation between the two eastern regions (Transylvania and Bukovina). This suggests that their genetic composition is specific to a certain degree. Moreover, populations of these two regions use different host ant species (Tartally et al. 2008; 2019) implying a certain level of ecological differentiation between them. Funk et al. (2012) defined the functional conservation units (FCU) as groups of populations with specific genetic composition which contribute significantly to the genetic diversity of an ESU. Based on this definition, we can assume that the populations of Transylvania and Bukovina belong to two different FCUs within the eastern ESU of P. nausithous (probably within the P. nausithous kijevensis subspecies).
Conclusions for future biology
Keeping in mind the significance of nature conservation nowadays, the study of genetic variation in natural populations seems to be an everlasting research field. While the aims of such research works are hardly changed, the methods to fulfil them are developing fast. New DNA techniques together with evolving statistical procedures provide more and more efficient tools to obtain deeper insight in the genetic composition and the past history of the species. Currently, the microsatellites are the most widespread markers in these studies. Nevertheless, a new technique, the analysis of SNPs (single nucleotide polymorphisms) via sequencing longer stretches of DNA in several individuals of the populations, is also expanding.
References
Allendorf FW, Luikart G (2009) Conservation and the genetics of populations. John Wiley & Sons
Árnyas E, Bereczki J, Tóth A et al (2006) Egg-laying preferences of the xerophilous ecotype of Maculinea alcon (lepidoptera: lycaenidae) in the aggtelek national park. Eur J Entomol 103:587–595
Bereczki J, Pecsenye K, Peregovits L et al (2005) Pattern of genetic differentiation in the Maculinea alcon species group (lepidoptera, lycaenidae) in Central Europe. J Zool Syst Evol Res 43:157–165
Bereczki J, Rácz R, Varga Z, Tóth JP (2015) Controversial patterns of Wolbachia infestation in the social parasitic Maculinea butterflies (Lepidoptera: Lycaenidae). Org Divers Evol 15:591–607
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet 19:233–257
Crandall KA, Bininda-Emonds ORP, Mace GM et al (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol 15:290–295
Earl DA, VonHoldt BM (2011) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Elmes GW, Thomas JA, Wardlaw JC (1991) Larvae of Maculinea rebeli, a large-blue butterfly, and their Myrmica host ants: wild adoption and behaviour in ant-nests. J Zool 223:447–460
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Figurny-Puchalska E, Gadeberg RME, Boomsma JJ (2000) Comparison of genetic population structure of the large blue butterflies Maculinea nausithous and M. teleius. Biodivers Conserv 9:419–432
Francis B, Green M, Payne C (1994) GLIM 4. The statistical system for generalised linear interactive modelling. Oxford University Press, New York
Funk WC, McKay JK, Hohenlohe PA et al (2012) Harnessing genomics for delineating conservation units. Trends Ecol Evol 27:489–496
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Hammer Ø, Harper DAT, Ryan PD (2001) Past: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Hollós A, Pecsenye K, Bereczki J et al (2012) Pattern of genetic and morphometric differentiation in Maculinea nausithous (Lepidoptera: Lycaenidae) in the Carpathian Basin. Acta Zool Acad Sci Hung 58:87–103
Hughes JB, Daily GC, Ehrlich PR (1997) Population diversity: its extent and extinction. Science 278(5338):689–692
Kizic M, Borovac M (2001) IMAGEJ: Image processing and analysis in java. http://rsb.info.nih.gov/ij/docs/pdfs/imagej-gui.pdf
Lukhtanov VA, Lukhtanov AG (1994) Die tagfalter nordwestasiens (Lepidoptera, Diurna). Herbipoliana 3:1–440
Maes D, Vanreusel W, Talloen W et al (2004) Functional conservation units for the endangered alcon blue butterfly Maculinea alcon in Belgium (Lepidoptera: Lycaenidae). Biol Cons 120:229–241
Moritz C (1994) Defining ‘evolutionarily significant units’ for conservation. Trends Ecol Evol 9:373–375
Munguira ML, Martin J (1999) Action plan for maculinea butterflies in Europe. Council of Europe, Strasbourg
Palsbøll PJ, Bérubé M, Allendorf FW (2007) Identification of management units using population genetic data. Trends Ecol Evol 22:11–16
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pecsenye K, Bereczki J, Tihanyi B et al (2007) Genetic differentiation among the Maculinea species (Lepidoptera: Lycaenidae) in eastern Central Europe. Biol J Lin Soc 91:11–21
Pecsenye K, Bereczki J, Juhász E et al (2015) Contrasting genetic structure in cuckoo and predatory Maculinea butterflies. Conserv Genet 16:939–954
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2010) Documentation for structure software: version 2.3 University of Chicago, Chicago
Rákosy L, Tartally A, Goia M et al (2010) The Dusky large blue—Maculinea nausithous kijevensis (Sheljuzhko, 1928) in the Transylvanian basin: new data on taxonomy and ecology. Nota Lepidopterol 33:31–37
Ritter S, Michalski SG, Settele J et al (2013) Wolbachia infections mimic cryptic speciation in two parasitic butterfly species, Phengaris teleius and P. nausithous (Lepidoptera: Lycaenidae). PLoS ONE 8:e78107
Ryder OA (1986) Species conservation and systematics: the dilemma of subspecies. Trends Ecol Evol 1:9–10
Schneider S, Roessli D, Excoffier L (2000) ver. 2000: A software for population genetic data analysis. Arlequin. Genetics and biometry laboratory, University of Geneva, Switzerland
Sheljuzhko L (1928) Neue Palaearktische Lycaenide. Lepidopterol Rundsch 2:44–46
Sneath PH, Sokal RR (1973) Numerical taxonomy. The principles and practice of numerical classification. W. H. Freeman, San Francisco
Sokal RR, Rohlf FJ (1981) Biometry. W.H. Freemann, San Francisco
Tartally A, Rákosy L, Vizauer TC, Goia M, Varga Z (2008) Maculinea nausithous exploits Myrmica scabrinodis in Transylvania: unusual host ant species of a myrmecophilous butterfly in an isolated region. Sociobiology 51(2):373–380
Tartally A, Thomas JA, Anton C et al (2019) Patterns of host use by brood parasitic Maculinea butterflies across Europe. Phil Trans R Soc B 374:20180202. https://doi.org/10.1098/rstb.2018.0202
Thomas JA (1984) The behaviour and habitat requirements of Maculinea nausithous (the dusky large blue butterfly) and M. teleius (the scarce large blue) in France. Biol Cons 28:325–347
Thomas JA, Elmes GW (2001) Food–plant niche selection rather than the presence of ant nests explains oviposition patterns in the myrmecophilous butterfly genus Maculinea. Proc R Soc Lond B: Biol Sci 268:471–477
Thomas JA, Wardlaw JC (1992) The capacity of a Myrmica ant nest to support a predacious species of Maculinea butterfly. Oecologia 91:101–109
Thomas JA, Elmes GW, Wardlaw JC et al (1989) Host specificity among Maculinea butterflies in Myrmica ant nests. Oecologia 79:452–457
Tuzov VK (1997) Guide to the butterflies of Russia and adjacent territories. Pensoft, Sofia
Vila M, Lundhagen AC, Thuman KA et al (2006) A new conservation unit in the butterfly Erebia triaria (Nymphalidae) as revealed by nuclear and mitochondrial markers. Ann Zool Fenn 43:72–79
Weir BS (1996) Genetic data analysis II: methods for discrete population genetic data. Sinauer Assoc. Inc, Sunderland
Wynhoff I (1998) Review: the recent distribution of the European Maculinea species. J Insect Conserv 2:15–27
Acknowledgements
The study was supported by the MacMan EVK2-CT-2001-00126 project. The authors are grateful to J. V. Sipos and Z. Ilonczay who effectively contributed to the sampling at several sites. The technical assistance of V. Mester in the electrophoretic work is very much respected. The support of the Nature Conservation Authorities of Hungary is highly appreciated.
Funding
Open access funding provided by University of Debrecen.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pecsenye, K., Bereczki, J., Hollós, A. et al. Evolutionary significant units (ESUs) and functional conservation units (FCUs) in the Phengaris (Maculinea) nausithous populations of East-Central Europe. BIOLOGIA FUTURA 74, 445–455 (2023). https://doi.org/10.1007/s42977-024-00211-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-024-00211-9