Abstract
Extracellular vesicles (EVs) are membrane-enclosed subcellular structures released by all cell types. EVs have important roles in both cellular homeostasis and intercellular communication. Recent progress in the field revealed substantial heterogeneity of EVs even within the size-based EV categories. Here we addressed the question whether the exportin-1 (XPO1)-mediated nuclear export of RNAs contributed to the EV heterogeneity. Size-based populations were separated from the conditioned media of three cell lines (U937, THP-1 and 5/4E8) in steady-state condition. The effects of activation and leptomycin B treatment (to inhibit the XPO1-mediated nuclear export of RNAs) were also tested in the case of the two monocytic cell lines. Agilent Pico and Small chips were used to characterize RNAs, fragment analysis was performed, and EV-associated miRNAs were tested by Taqman assays. As expected, we found the highest small RNA/total RNA ratio and the lowest rRNA/total RNA proportion in small EVs (~ 50–150 nm). Profiles of the small RNAs within different size-based EV categories significantly differed based on the activation status of the EV releasing cells. Leptomycin B had a differential inhibition on the tested small RNAs in EVs, even within the same EV size category. A similar heterogeneity of the EV miRNA content was observed upon cellular activation and nuclear export inhibition. Here we complement the already existing knowledge on EV heterogeneity by providing evidence that the RNA cargo varies depending on the EV size-based category, the releasing cell type, the functional status of the releasing cells and the exportin-1-mediated nuclear export of RNAs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Extracellular vesicles (EVs) are released by cells to maintain cellular homeostasis and to promote intercellular communication with neighbouring or with distant cells by delivering molecular cargoes. Cells can secrete different types of EVs, which can be distinguished by their size, morphology and biogenesis (György et al. 2011; Mathieu et al. 2019). Two basic types of EVs can be distinguished on the basis of their biogenesis, namely the endosome-derived exosomes and the plasma membrane-derived EVs (ectosomes). However, clear molecular markers for the different biogenetic pathways are not available (Buzás 2022). In general, ESCRT proteins and their associated accessory proteins (Alix, TSG101, HSC70 and HSP90β) are usually present in exosomes, as exosome formation and multivesicular body (MVB) transport are regulated by ESCRT proteins (Trajkovic et al. 2008). CD9, CD63 and CD81 are proteins of the tetraspanin family. These transmembrane proteins and other plasma membrane associated proteins are commonly found in EVs (Turiák et al. 2011). Unlike exosomes and MVs, large EVs such as apoptotic bodies may even contain intact organelles and chromatin and small amounts. The proteomic profiles of cell lysates and apoptotic bodies are quite similar, whereas they differ significantly from exosomes (Théry et al. 2001; Crescitelli et al. 2013). Since there are no clear markers for EVs, large EVs (lEVs), medium-size (mEVs) and small EVs (sEVs) are operational categories suggested by the MISEV2018 guidelines until established molecular markers, specific for the different biogenetic routes become available (Théry et al. 2018).
The cargo composition of EVs is dependent on the donor cells (Colombo et al. 2014; Valadi et al. 2007). EVs selectively carry cell-specific proteins, nucleic acids, metabolites, glycans and lipids, all contributing to the cargo signature.
Numerous studies demonstrated diversity of EVs based on their proteomic, lipidomic or metabolite composition. RNAs attracted an outstanding attention in the past years. The mechanisms regulating the loading of RNAs into EVs for secretion are under intense investigation. Studies have focused on RNA-binding proteins (RBPs) for RNA loading into sEVs (Villarroya-Beltri et al. 2013; Li et al. 2004). sEVs and miRNAs have been reported to have both non-selective secretion (Tosar et al. 2015) and true selective or preferential sorting mechanisms in various studies (Teng et al. 2017; Temoche-Diaz et al. 2019).
Here we investigated whether inhibition of exportin-1 (XPO1), a molecule heavily involved in the nuclear export of RNAs, had an impact on the amount and composition of RNAs of EVs secreted under steady-state condition and upon activation. Our data provide evidence for the differential involvement of exportin-1-mediated RNA nuclear export in determining the RNA content of cell-derived EVs.
Materials and methods
Cell cultures
U937 human histiocytic lymphoma cell line, THP-1 (TIB-202) human acute monocytic lymphoma cell lines were obtained from ATCC (Manassas, VA) and were cultured in RPMI 1640 medium (Sigma). The 5/4E8 Th1 T hybridoma cell line, generated by our group (Buzás et al. 2003), was cultured in DMEM. All media contained 10% FBS (Gibco, BioSera), 1% Antibiotic Antimycotic Solution for Cell Culture (EMD Millipore) and 1% glutamine (Gibco).
Extracellular vesicle separation
Before EV isolation, cells were transferred to a CELLine bioreactor (0.5–2.0 × 108/15 mL concentration, Integra Biosciences, Chur, Switzerland) in 450 mL of cell type-matched medium containing 2.5% EV-free FBS. The FBS was centrifuged by 2,000 g 20 min, and by 120,000 g 16 h, after which it was filtered through a 0.1-μm filter. In a few experiments, U937 cells were activated with 1 mg/mL lipopolysaccharide (LPS, Sigma-Aldrich). THP-1 cells were stimulated with 25 ng/mL Phorbol 12-Myristate 13-acetate (PMA, Sigma-Aldrich). For both cell types, 10 nM leptomycin B (LMB, Sigma-Aldrich) was used to inhibit the nuclear export of RNAs. Cells were cultured for 24 h under the above conditions.
Three different size-based EV subpopulations were separated including lEVs, mEVs and sEVs by the combination of gravity-driven size filtration and differential (ultra)centrifugation. Briefly, cells were removed by centrifugation (2 × 200 g for 5 min). Any remaining cells were removed by gravity-driven filtration through a 5-μm filter (Millipore, Billerica, MA). Next, 2,000 g centrifugation was used for 20 min at room temperature to pellet lEVs. The supernatant was filtered by gravity through a 0.8-μm filter (Millipore), and the filtrate was centrifuged at 12,500 g for 20 min at room temperature to pellet mEVs. Finally, the supernatant was filtered through a 0.2-μm filter and was ultracentrifuged in an Optima MAX-XP bench top ultracentrifuge with MLA-55 rotor (Beckman Coulter Inc., Brea, CA) at 100,000 g for 70 min at 4 °C to pellet sEVs. Each EV pellet was resuspended once in PBS and re-centrifuged under the same condition with an MLS-50 rotor that was originally used for pelleting (Supplementary Figure 1).
Western blot
For the assay, after washing of the cells and the EV pellets, samples were lysed in lysis buffer (CellLytic; Sigma-Aldrich) containing protease inhibitor (cOmplete Protease Inhibitor Coctail; Roche). For cells and lEVs, the exact protein concentration was determined by microBCA (Thermo Scientific). For mEVs and sEVs, the pellet was resuspended in 60 μL lysis buffer. For cells, 20 μg/lane, for lEVs, 10 μg/lane of protein and for mEVs and sEVs, 12 μL resuspended pellet/lane were used. For sample preparation prior to electrophoresis, we used a fivefold concentrated Laemmli buffer and denatured the proteins at 95 °C for 5 min and then applied 15 µL to the gel (CriterionTMTGX™ Precast Gels 8–16%, 26 well; Bio-Rad, we used Spectra™ Multicolor Broad Range Protein Ladder, Thermo Scientific). Electrophoresis was carried out at a constant voltage of 130 V for 1.5 h, and the gel ran while the tank was on ice. The proteins were then blotted onto polyvinylidene fluoride (PVDF, Bio-Rad) membranes (for 2.5 h) using transfer buffer (25 mM Tris, 192 mM glycine, 20 v/v% methanol; pH 8.3). The membranes were then blocked with a SuperBlock (TBS) Blocking Buffer (Thermo Scientific) solution at room temperature for 2 h. They were then incubated with rabbit primary antibodies reactive to EV marker proteins (Supplemetary Table1) overnight at 4 °C, followed by goat polyclonal anti-rabbit HRP-labelled secondary antibody (Abcam) with an incubation time of 30 min. The chemiluminescent signal was detected using Clarity MaxWestern ECL Substrate (Bio-Rad). Detection was performed using Imager Chemi Premium (VWR).
Tunable resistive pulse sensing (tRPS) measurements
Tunable resistive pulse sensing is a technique used in order to determine particle size and concentration (Maas et al. 2014). EV preparations were subjected to tRPS analysis using a qNano instrument (IZON Science, New Zealand). Serial dilutions of each EV preparation were subjected to particle number analysis for 5 min using 5 mbar pressure and NP200, NP400 and NP2000 nanopore membranes stretched between 45 and 47 mm. Calibration was performed using known concentration of beads CPC100B, or CPC400E and CPC2000C (all from IZON) diluted 1:1,000 in 0.2-μm filtered PBS.
Transmission electron microscopy (TEM)
TEM is a common electron microscopy method used to evaluate the morphology of EVs. After centrifugation, the supernatants were carefully removed and the pellets were fixed in RT for 60 min in fixative (2% glutaraldehyde and 2% PFA (Sigma) in PBS), followed by fresh fixative overnight (4̊C). The next day, after washing twice with PBS, the preparations were postfixed in 1% OsO4 (Taab, Aldermaston, Berks, UK) for 30 min. After rinsing with distilled water, pellets were dehydrated in graded ethanol, including block staining with 1% uranyl acetate in 50% ethanol for 30 min, and embedded in Taab 812 (Taab). After overnight polymerization at 60 °C and sectioning for EM, the ultrathin sections were analysed using a HITACHI 7100 electron microscope equipped with a Megaview II (lower resolution, Soft Imaging System, Munster, Germany) digital camera.
RNA isolation and detection
Cells and EVs were lysed in 700 µl QIAzol lysis buffer, and RNA was isolated using RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). RNA concentration was measured using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). RNA samples profiles were further assessed on an RNA 6000 Pico chip and a Small chip using the 2100 Electrophoresis Bioanalyzer (Agilent Technologies, USA).
miRNA detection
Based on 10 ng total RNA, TaqMan™ Advanced miRNA cDNA Synthesis Kit was used for ligation-based universal reverse transcription for a streamlined and highly sensitive workflow (Applied Biosystems, USA). GeneAmp™ PCR System 9700 was used. The RT product was used in a further tenfold dilution; qPCR was performed using SensiFAST™ Probe No-ROX Kit (Bioline, Taunton, MA, USA) and Advanced miRNA Assays (Life Technologies) on a BioRad CFX384 Touch Real-Time PCR machine (run mode: Enzyme activation 95 °C 2 min, Cycle: 95 °C 5 s and 60 °C 30 s 40×). Data were collected at the end of each cycle. Any samples with Ct values > 35 we considered to be below our detection limit and we excluded them. The specific Taqman miRNA assays used in this study are summarised in Supplementary Table 2. Normalized relative expression levels were calculated using 2−ΔΔCT. Normalization was performed using a stably expressed reference miR (miR-425) and was validated using NormFinder's module (Supplementary Table 3).
Results
Representation of different RNA regions in cell lines and their secreted vesicles
We separated sEVs, mEVs and lEVs from the conditioned medium of the THP1and U937monocyte cell lines and that of the 5/4E8 Th1 T cell hybridoma. Total RNA was extracted from the cells and the different size-based EV categories and were subjected to fragment analysis using a Bionalyzer Pico chip. Figure 1A shows representative electropherograms obtained for U937 cells and the U937-derived EVs. These RNAs correspond to the different RNA regions of the recorded Bioanalyzer electrophoregrams the percentage distribution of which is shown in Fig. 1B. The “Small RNA region” was overrepresented in the sEVs compared to the other two size-based EV categories, and the difference was even more striking when compared to the RNA profile of the cells. The other striking finding was that while cells, mEVs and lEVs contained significant amounts of 18S and 28S ribosomal RNA molecules, the 18S and 28S ribosomal RNAs was barely found in sEVs. While the ratio of 28S:18S RNAs in cells was 2:1, this ratio was close to 1:1 in lEVs and mEVs. These data were in line what we found in the case of the two other cell lines (THP1 and 5/4E8) (Fig. 1C).
Characterization of total RNA with Bioanalyzer. A Representative image showing the total RNA content of U937 cells and their secreted extracellular vesicles (lEV, mEV, sEV) obtained using the RNA 6000 PicoChip Kit (Agilent Technologies, USA). Characterization of total RNA with Bioanalyzer. The electropherograms show the size distribution in nucleotides (nt) and fluorescence intensity (FU) of total RNA. The first peak at 25 nt is an internal standard. B Distribution of RNA regions identified in total RNA electrophorogram analysis of steady-state U937 cells (mean ± SD; n = 4). C Ratio of small RNAs and rRNAs and the total RNA, and the proportion of 28S and 18S ribosomal RNAs (ANOVA, *p < 0.05; ***p < 0.001) in cells and EVs
Distribution of different length RNAs within the small region in cell lines and their released EVs
Our next question concerned the distribution of the different length RNAs within the cells and the secreted EVs. U937 cells and the released vesicles were compared in steady-state condition, upon exposure to LPS and upon exposure to the nuclear export (exportin 1)-inhibitor leptomycin B (LMB).
Here we analysed the RNA samples by a Bioanalyzer Small chip. Figure 2A shows representative electropherograms of U937 cells and the secreted EVs. Figure 2B demonstrates the distribution of the fragment lengths expressed as ranges of nucleotide (nt) numbers. The regions corresponding to different small RNA species were compared. Within the small RNAs (14–150 nt), miRNAs (14–30 nt), tRNAs (50–80 nt), 5S rRNAs (80–100 nt), and 5.8S rRNAs (130–150 nt) were distinguished.
Analysis of small RNA regions. Electrophoretic profiles of small RNAs obtained using the Small RNA Assay on the Bioanalyzer (Agilent Technologies, USA) from U937 cells and their secreted lEVs, mEVs and sEVs. A The electropherograms show the size distribution in nucleotides (nt) and fluorescence intensity (FU) of small RNAs. The peak at 4 nt is an internal standard. The red line indicates 1 mg/mL LPS treatment, the blue line indicates the steady-state condition and the green line shows 10 nM LMB treatment. B The regions corresponding to the different small RNA species (miRNAs 14–30 nt, tRNAs 50–80 nt, rRNAs 5S 80–100 nt, 5.8S 130–150 nt). Pie charts of the small RNA species analysed and their percentages in small RNAs (14–150 nt). Mean values determined in steady-state condition (control, n = 4), and upon the two different treatment types (n = 3 for both treatments) are shown. C Ratios of different RNA types determined in steady-state condition of U937, THP1 and 5/4E8 cells
In our analysis, in steady state of U937 cells, lEVs and mEVs, the RNAs of 50–80 nt were the most abundant RNA species. The 5S and 5.8S ribosomal RNAs were significantly lower in the sEVs as compared to cells (p < 0.0001, t test). Similarly, 5S and 5.8S RNAs were significantly lower in sEVs as compared to lEVs (p = 0.0004, t test). This is similar to what was seen when using an Agilent Pico chip (18S and 28S). Some of the RNAs in the 30–50 nt region are considered to contain tRNA fragments (tRFs) (Weng et al. 2022; Kazimierczyk et al. 2022). Using a Small chip, we found that the ratio of rRNAs (80–100 nt + 130–150 nt regions)/tRFs (30–50 nt) + tRNAs (50–80 nt region) was 1.2 ± 0.12 in cells, 1.2 ± 0.23 in lEVs, 0.5 ± 0.13 in mEVs, and 0.3 ± 0.07 in sEVs. These ratios were also examined in the case of THP-1 and 5/4E8 cell lines, and showed a similar trend. In the case of lEVs, this ratio was somewhat lower in lEVs of THP-1 and 5/4E8 cells as compared to those released by U937 cells (0.8 ± 0.12 and 0.7 ± 0.15, respectively). (Fig. 2C). While U937 cells in steady state expressed a low percentage of microRNAs (miRNAs), the amount of miRNAs was substantially higher in EVs, especially in sEVs, where they accounted for at least 20% of all small RNAs. LPS stimulation further increased the amount of 14–30 nt RNAs (which is considered to contain most of the miRNAs) in sEVs (p = 0.0264, t test). In contrast, LMB did not induce any significant change in the proportion of miRNAs in sEVs (p = 0.1243, t test) (Fig. 2B).
The presence of miRNAs in cell lines and their released EVs
We also investigated whether miRNAs in different size-based EV subpopulations differed in steady-state conditions, or when U937 and THP1 cells were stimulated with LPS and PMA, respectively. Supplementary Figure 3 summarizes the data obtained from the analysis of cells and the released mEVs and sEVs. Both LPS/PMA and LMB exposure resulted in multiple changes in miRNA expression both in cells and in the secreted EVs. As shown in Fig. 3, different patterns were observed. To analyse the effect of LMB treatment, we wanted to see how the miRNA levels of mEVs and sEVs differed when compared to the cells in the case of both monocytic cell lines (U937 and THP1). Category 1: no difference cell-mEV, no difference cell-sEV: let-7g, miR-16; category 2: there is a difference cell-mEV, no difference cell-sEV: miR-148; category 3: no difference cell-mEV, there is a difference cell-sEV: let-7a; and category 4: there is difference cell-mEV, there is difference cell-sEV: miR-26. Next, we also examined a 5th category where mEV and sEV were compared. Here we found three miRNAs (miR-126, miR-148, miR-423) that showed a difference between EVs in both cell line-derived EVs. It is also interesting that not only a quantitative difference was observed between miR-148 and miR-423, but also that opposite change of expression was observed in response to LMB (as compared to the untreated control) in mEVs and cells or mEVs and sEVs.
Comparative analysis of the relative expression of miRNAs in cells and EVs. Statistical comparison the 2−ΔΔCT values was carried out by ANOVA. The fold change (FC) values show gene expressions relative to values obtained in steady-state condition. The p-values are shown in the heat map. A U937 cells stimulated by LPS, B inhibition of U937 cells by LMB, C THP1 cells stimulated by PMA, D THP1 cells treated by LMB. The first columns show the comparison between cells and mEVs, and the second columns show the comparison between cells and sEVs, while the third column shows the comparison between mEVs and sEVs. The colours indicate significance values: blue is non-significant and red is highly significant. In the case of the mEVs of THP1 cells, miR-484 was not detectable (indicated by crossed boxes). In steady-state condition (control, n = 4), and upon the two different treatment types (n = 3 for both treatments) are shown
These data suggest that the reported EV heterogeneity within size-based EV subpopulations is also reflected by different miRNA patterns of EVs.
Discussion
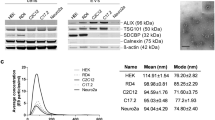
The exponentially growing scientific interest attracted by EVs can be largely attributed to the RNA content of EVs. In this study, we separated different size-based subpopulations of EVs derived from the conditioned media of cells. We confirmed the EV nature of the tested by particles by TRPS, transmission electron microscopy and Western blot. Our Western blot data confirmed that the known sEV-related molecules (Alix, CD63, flotillin-1 and TSG101) were indeed present in our small EV preparations. Since the cells produce different amounts of these proteins, it was not surprising that this difference was also observed in the EVs themselves.
Several types of RNAs have been identified in EVs including mRNAs (Valadi et al. 2007) and different non-coding RNAs (e.g. ncRNAs such as transfer RNAs, Y RNAs, vault RNAs, small nuclear and nucleolar RNAs, rRNAs, lncRNAs and PIWI-interacting RNAs) (O'Brien et al. 2020). From among the many important features of EV-associated RNAs, we have to emphasize the transfer of functional RNAs to recipient cells supported by a plethora of evidences (Hill et al. 2013; Mateescu et al. 2017).
In our work, we observed a clear enrichment of tRNAs and tRFs in mEVs and sEVs as compared to rRNAs, which is consistent with other published reports (Temoche-Diaz et al. 2019; Tosar et al. 2020). In line with our earlier findings, here we found a preferential enrichment of small RNAs in sEVs (Crescitelli et al. 2013). This is particularly important given that sEVs can efficiently cross barriers (such as the blood brain barrier or the blood testis barrier). The proposed distinguished role of sEVs in intercellular communication is in line with the small size and small RNA content of these vesicles.
We found differential expression of miRNAs in cells and in EVs. The tested miRNAs were selected based on their known expression in monocytes/macrophages. Among them, miR-223 is one of the best characterized miRNAs in monocytes/macrophages, while in EVs, miR-16 and members of the let-7 family are abundantly characterized. It was shown earlier that during LPS- or PMA-induced monocyte differentiation, there was a decrease in miR-223 along with a reduced expression of miR-15 and miR-16 (Li et al. 2010). In our experiments, there was no change in the expression of miR-223 és miR-16 in U937 cells upon LPS treatment. In contrast, in mEVs and sEVs of LPS-treated cells, there was a significant decrease in miR-16 compared to the releasing LPS-treated cells, but there was no difference compared to the EVs of untreated cells. In the case of miR-15, there was no LPS-induced change either in the cells or in the EVs. Upon treatment with PMA, miR-223 and miR-16 were unchanged in THP1 cells and released EVs, while miR-15 was significantly reduced in sEVs.
A possible explanation for the lack or reduction of miR-223 upon LPS or PMA stimuli in our experiments is that macrophage differentiation takes several days, while in our experiments, we exposed our cells to 24 h stimulation. Our data suggest that EV-associated miR-16 or miR-15 may react to the differentiation stimuli earlier than the EV-releasing monocytic cells.
In the present study, we addressed a previously unexplored or hardly explored aspect of the EV RNA biology, the involvement of XPO1-mediated nuclear export of RNAs in determining the RNA cargo of EVs.
Nuclear export is a highly regulated process that occurs through the nuclear pores (Cullen 2003). In humans, there are seven exportin molecules playing key roles in the nuclear export of proteins, tRNAs, and miRNAs. They recognize the nuclear export signal (NES) on proteins of ribonucleoprotein complexes associated with RAN-GTP, to translocate them to the cytoplasm. While some of the exportin molecules only recognize a low number of proteins or miRNAs, XPO1 can bind to NES of more than 200 proteins and miRNAs. XPO1 is universally expressed by human cells suggesting a very important role of this protein in cellular homeostasis. Given the ubiquitous role of EVs in regulating intercellular communication, which is largely based on the EV cargo, here we aimed at investigating the question whether blocking XPO1 has an impact on loading of the RNA cargo to different types of EVs.
Our data show that LMB treatment significantly and differentially altered the RNA profiles of cells and EVs. We observed different miRNA expression patterns in cells and EVs. In the case of miR-126 and miR-484, an LMB-induced inhibition was visible in U937 cells, whereas no inhibition was observed in THP1 cells. Interestingly, we found three miRNAs (miR-126, miR-148, miR-423) that showed a difference between mEVs and sEVs in both monocytic cell lines. In addition, two of the above miRNAs changed discordantly upon LMB treatment. This may suggest differential role of XPO1 in determining the miRNA content of EV subtypes.
Currently, significant research efforts are devoted worldwide to understand and to interfere with EV RNA-mediated effects on cells both in vitro and in vivo.
Our study did not investigate whether exportin-1 inhibition could suppress EV RNA-mediated pathological processes. However, the observation that LMB treatment significantly and differentially altered the RNA profiles of EVs may suggest that LMB treatment could possibly hold therapeutic potential in certain pathological conditions.
References
Buzás EI (2022) The roles of extracellular vesicles in the immune system. Nat Rev Immunol. https://doi.org/10.1038/s41577-022-00763-8
Buzás EI, Hanyecz A, Murad Y, Hudecz F, Rajnavölgyi E, Mikecz K, Glant TT (2003) Differential recognition of altered peptide ligands distinguishes two functionally discordant (arthritogenic and nonarthritogenic) autoreactive T cell hybridoma clones. J Immunol 171:3025–3033. https://doi.org/10.4049/jimmunol.171.6.3025
Colombo M, Raposo G, Théry C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. https://doi.org/10.1146/annurev-cellbio-101512-122326
Crescitelli R, Lässer C, Szabó TG, Kittel Á, Eldh M, Dianzani I, Buzás EI, Lötvall JO (2013) Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. https://doi.org/10.3402/jev.v2i0.20677
Cullen BR (2003) Nuclear RNA export. J Cell Sci 116(4):587–597. https://doi.org/10.1242/jcs.00268
György B, Szabó TG, Pásztói M, Pál ZS, Misják P, Aradi B, László V, Pállinger É, Pap E, Kittel Á, Nagy G, Falus A, Buzás EI (2011) Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 68:2667–2688. https://doi.org/10.1007/s00018-011-0689-3
Hill AF, Pegtel DM, Lambertz U, Leonardi T, O’Driscoll L, Pluchino S, Ter-Ovanesyan D, Nolte-’t Hoen EN (2013) ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J Extracell Vesicles. https://doi.org/10.3402/jev.v2i0.22859
Kazimierczyk M, Wojnicka M, Biała E, Żydowicz-Machtel P, Imiołczyk B, Ostrowski T, Kurzyńska-Kokorniak A, Wrzesinski J (2022) Characteristics of transfer RNA-derived fragments expressed during human renal cell development: the role of Dicer in tRF biogenesis. Int J Mol Sci 23(7):3644. https://doi.org/10.3390/ijms23073644
Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, Stadtman ER, Yang DC, Chock PB (2004) Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc Natl Acad Sci 101:8551–8556. https://doi.org/10.1073/pnas.0402889101
Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG (2010) MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 11(9):799–805. https://doi.org/10.1038/ni.1918
Maas SL, De Vrij J, Broekman ML (2014) Quantification and size-profiling of extracellular vesicles using tunable resistive pulse sensing. J vis Exp. https://doi.org/10.3791/51623
Mateescu B, Kowal EJ, Balkom BW, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FW, Das S, Driedonks TA et al (2017) Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—an ISEV position paper. J Extracell Vesicles 6(1):1286095. https://doi.org/10.1080/20013078.2017.1286095
Mathieu M, Martin-Jaular L, Lavieu G, Théry C (2019) Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21:9–17. https://doi.org/10.1038/s41556-018-0250-9
O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO (2020) RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol 21(10):585–606. https://doi.org/10.1038/s41580-020-0251-y
Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, Yao J, Fadadu RP, Lambowitz AM, Schekman R (2019) Distinct mechanisms of microrna sorting into cancer cell-derived extracellular vesicle subtypes. Elife 8:e47544. https://doi.org/10.7554/eLife.47544
Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML, Yan J, Miller DM, Zhang HG (2017) MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 8:14448. https://doi.org/10.1038/ncomms14448
Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S (2001) Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol 166(12):7309–7318. https://doi.org/10.4049/jimmunol.166.12.7309
Théry C, Witwer KW et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7(1):1535750. https://doi.org/10.1080/20013078.2018.1535750
Tosar JP, Gámbaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A (2015) Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucl Acids Res 43:5601–5616. https://doi.org/10.1093/nar/gkv432
Tosar JP, Segovia M, Castellano M, Gámbaro F, Akiyama Y, Fagúndez P, Olivera Á, Costa B, Possi T, Hill M, Ivanov P, Cayota A (2020) Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Res 48(22):12874–12888. https://doi.org/10.1093/nar/gkaa674
Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P et al (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319(5867):1244–1247. https://doi.org/10.1126/science.1153124
Turiák L, Misják P, Szabó TG, Aradi B, Pálóczi K, Ozohanics O, Drahos L, Kittel Á, Falus A, Buzás EI, Vékey K (2011) Proteomic characterization of thymocyte-derived microvesicles and apoptotic bodies in BALB/c mice. J Proteom 74(10):2025–2033. https://doi.org/10.1016/j.jprot.2011.05.023
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. https://doi.org/10.1038/ncb1596
Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. https://doi.org/10.1038/ncomms3980
Weng Q, Wang Y, Xie Y, Yu X, Zhang S, Ge J, Li Z, Ye G, Guo J (2022) Extracellular vesicles-associated tRNA-derived fragments (tRFs): biogenesis, biological functions, and their role as potential biomarkers in human diseases. J Mol Med (berl) 100(5):679–695. https://doi.org/10.1007/s00109-022-02189-0
Acknowledgements
This work was supported by National Research, Development and Innovation Office NKFIH, Hungary, NVKP_16-1-2016-0017, the Hungarian Scientific Research Fund (OTKA K120237), VEKOP-2.3.2-16-2016-000002, VEKOP-2.3.3-15-2017-00016, H2020-MSCA-ITN-2017-722148 TRAIN EV and Higher Education Excellence Program (FIKP) Therapeutic Thematic Programme. This study was supported by the Hungarian Thematic Excellence Programme No. TKP2021-EGA-23 and the National Cardiovascular Laboratory Program RRF-2.3.1-21-2022-00003. The project has received funding from the EU’s Horizon 2020 Research and Innovation Programme under grant agreement No. 739593.
Funding
Open access funding provided by Semmelweis University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
E.I.B. is a member of the Advisory Board of Sphere Gene Therapeutics Inc. (Boston, MA, USA) and ReNeuron—Cell and Gene Therapy (UK).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pálóczi, K., Buzas, E.I. & Falus, A. Differential impact of exportin-1-mediated nuclear export of RNAs on the RNA content of extracellular vesicle subpopulations. BIOLOGIA FUTURA 74, 91–99 (2023). https://doi.org/10.1007/s42977-023-00157-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-023-00157-4