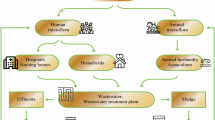
Abstract
Antibiotic resistance bacteria (ARB) and antibiotic resistance genes (ARGs) are now considered major global threats. The Kongsfjorden and Krossfjorden are the interlinked fjords in the Arctic that are currently experiencing the effects of climate change and receiving input of pollutants from distant and regional sources. The present study focused on understanding the prevalence of antibiotic resistance of retrievable heterotrophic bacteria from the sediments of adjacent Arctic fjords Kongsfjorden and Krossfjorden. A total of 237 bacterial isolates were tested against 16 different antibiotics. The higher resistance observed towards Extended Spectrum β-lactam antibiotic (ESBL) includes ceftazidime (45.56%) followed by trimethoprim (27%) and sulphamethizole (24.05%). The extent of resistance was meagre against tetracycline (2.53%) and gentamycin (2.95%). The 16S rRNA sequencing analysis identified that Proteobacteria (56%) were the dominant antibiotic resistant phyla, followed by Firmicutes (35%), Actinobacteria (8%) and Bacteroidetes. The dominant resistant bacterial isolates are Bacillus cereus (10%), followed by Alcaligenes faecalis (6.47%), Cytobacillus firmus (5.75%) Salinibacterium sp. (5%) and Marinobacter antarcticus (5%). Our study reveals the prevalence of antibiotic resistance showed significant differences in both the inner and outer fjords of Kongsfjorden and Krossfjorden (p < 0.05). This may be the input of antibiotic resistance bacteria released into the fjords from the preserved permafrost due to the melting of glaciers, horizontal gene transfer, and human influence in the Arctic region act as a selection pressure for the development and dissemination of more antibiotic resistant bacteria in Arctic fjords.





Similar content being viewed by others
References
Ahmad M, Khan AU (2019) Global economic impact of antibiotic resistance: a review. J Glob Antimicrob Resist 19:313–316
Holmes AH, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, Guerin PJ, Piddock LJV (2016) Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387(10014):176–187
Davies J, Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74(3):417–433
Pruden A, Pei R, Storteboom H, Carlson KH (2006) Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol 40(23):7445–7450
D’Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD (2011) Antibiotic resistance is ancient. Nature 477:457–461
Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259
Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen ALV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48(D1):D517–D525
Zhang T, Li J, Wang N, Wang H, Yu L (2022) Metagenomic analysis reveals microbiome and resistome in the seawater and sediments of Kongsfjorden (Svalbard, High Arctic). Sci Total Environ 809:151937
Hernández J, González-Acuña D (2016) Anthropogenic antibiotic resistance genes mobilization to the polar regions. Infect Ecol Epidemiol 6(1):32112
Scott LC, Lee N, Aw TG (2020) Antibiotic resistance in minimally human-impacted environments. Int J Environ Res Public Health 17(11):3939
Hatha AAM, Divya PS, Saramma AV, Rahiman M, Krishnan KP (2013) Migratory bird, Branta leucopis (barnacle goose), a potential carrier of diverse Escherichia coli serotypes into pristine Arctic environment. Curr Sci 104(8):1078–1080
Li J, Cao J, Zhu YG, Chen QL, Shen F, Wu Y, Xu S, Fan H, Da G, Huang RJ, Wang J, de Jesus AL, Morawaska L, Chan CK, Peccia J, Yao M (2018) Global survey of antibiotic resistance genes in air. Environ Sci Technol 52(19):10975–10984
Tan L, Li L, Ashbolt N, Wang X, Cui Y, Zhu X, Xu Y, Yang Y, Mao D, Luo Y (2018) Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci Total Environ 621:1176–1184
Perron GG, Whyte L, Turnbaugh PJ, Goordial J, Hanage WP, Dantas G, Desai MM (2015) Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS ONE 10(3):e0069533
McCann CM, Christgen B, Roberts JA, Su JQ, Arnold KE, Gray ND, Zhu YG, Graham DW (2019) Understanding drivers of antibiotic resistance genes in high Arctic soil ecosystems. Environ Int 125:497–504
Sjölund M, Bonnedahl J, Hernandez J, Bengtsson S, Cederbrant G, Pinhassi J, Kahlmeter G, Olsen B (2008) Dissemination of Multidrug-resistant Bacteria into the Arctic. Emerg Infect Dis 14(1):70–72
Middleton JH, Ambrose A (2005) Enumeration and antibiotic resistance pattern of fecal indicator organisms isolated from migratory Canada geese (Branta Canadensis). J Wildl Dis 41(2):334–341
Hatha AAM, Neethu CS, Nikhil SM, Rahiman KMM, Krishnan KP, Saramma AV (2015) Relatively high antibiotic resistance among heterotrophic bacteria from arctic fjord sediments than water–evidence towards better selection pressure in the fjord sediments. Polar Sci 9(4):382–388
Akhil Prakash E, Hromádková T, Jabir T, Vipindas PV, Krishnan KP, Mohamed Hatha AA, Briedis M (2022) Dissemination of multidrug resistant bacteria to the polar environment - role of the longest migratory bird Arctic tern (Sterna paradisaea). Sci Total Environ 815:152727
Colby GA, Ruuskanen MO, St. Pierre KA, St. Louis VL, Poulain AJ, Aris-Brosou S (2020) Warming climate is reducing the diversity of dominant microbes in the largest high Arctic lake. Front Microbiol 11:561194
Mogrovejo DC, Perini L, Gostinčar C, Sepčić K, Turk M, Ambrožič-Avguštin J, Brill FHH, Gunde-Cimerman N (2020) Prevalence of Antimicrobial Resistance and hemolytic phenotypes in Culturable Arctic Bacteria. Front Microbiol 11:570
Intergovernmental Panel on Climate Change [IPCC] (2007) Climate change 2007: impacts, adaptation and vulnerability. in Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. University, Cambridge
Livermore DM (2003) Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis 36:S11–S23
Altizer S, Ostfeld RS, Johnson PTJ, Kutz S, Harvell CD (2013) Climate change and infectious diseases: from evidence to a predictive framework. Science 341(6145):514–519
Tam HK, Wong CMVL, Yong ST, Blamey J, González M (2015) Multiple-antibiotic-resistant bacteria from the maritime Antarctic. Polar Biol 38:1129–1141
Vishnupriya S, Jabir T, Adarsh BM, Kattatheyil H, Shahana Kabeer S, Krishnan KP, Radhakrishnan CK, Mohamed Hatha AA (2023) Diversity of complex polysaccharide degrading bacteria from the sediments of interlinked high Arctic fjords, Svalbard. Reg Stud Mar Sci 63:102989
Wilson K (1997) Preparation of genomic DNA from bacteria. Current protocols in Molecular Biology, In: Ausubel FM, Brent R, Kingston RE, Moore DD., Seidman JG, Smith JA, Struhl, K (eds), 2.4.1–2.4.5
Thomas F, Sinha R, Krishnan KP (2020) Bacterial community structure of a glacio-marine system in the Arctic (Ny-Ålesund, Svalbard). Sci Total Environ 718:135264
Sinha RK, Krishnan KP, Kerkar S, Thresyamma DD (2017) Influence of glacial melt and Atlantic water on bacterioplankton community of Kongsfjorden, an Arctic fjord. Ecol Indic 82:143–151
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027
Yu G (2020) Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinform 69(1):e96
Wickham H (2006) An introduction to ggplot: An implementation of the grammar of graphics in R. Statisticcs:1–8
Bauer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496
Krumperman PH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46(1):165–170
R Core Team (2021) R: A language and environment for statistical computing. R foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Shariatinajafabadi M, Wang T, Skidmore AK, Toxopeus AG, Kölzsch A, Nolet BA, Exo KM, Griffin L, Stahl J, Cabot D (2014) Migratory herbivorous waterfowl track satellite-derived green wave index. PLoS ONE 9(9):e108331
Yuan M, Yu Y, Li HR, Dong N, Zhang XH (2014) Phylogenetic diversity and Biological Activity of Actinobacteria isolated from the Chukchi Shelf Marine sediments in the Arctic Ocean. Mar Drugs 12(3):1281–1297
Singh P, Singh SM, Singh RN, Naik S, Roy U, Srivastava A, Bölter M (2017) Bacterial communities in ancient permafrost profiles of Svalbard, Arctic. J Basic Microbiol 57(12):1018–1036
Van Goethem MW, Pierneef R, Bezuidt OKI, Van De Peer Y, Cowan DA, Makhalanyane TP (2018) A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6:40
Rabbia V, Bello-Toledo H, Jiménez S, Quezada M, Domínguez M, Vergara L, Gómez-Fuentes C, Calisto-Ulloa N, González-Acuña D, López J, González-Rocha G (2016) Antibiotic resistance in Escherichia coli strains isolated from Antarctic bird feces water from inside a wastewater treatment plant, and seawater samples collected in the Antarctic Treaty area. Polar Sci 10(2):123–131
Maiden MCJ (1998) Horizontal Genetic Exchange Evolution, and spread of antibiotic resistance in Bacteria. Clin Infect Dis 27:S12–S20
Glad T, Bernhardsen P, Nielsen KM, Brusetti L, Andersen M, Aars J, Sundset MA (2010) Bacterial diversity in faeces from polar bear (Ursus maritimus) in Arctic Svalbard. BMC Microbiol 10:1–10
Sudha A, Augustine N, Thomas S (2013) Emergence of multidrug resistant bacteria in the Arctic, 79ºN. J Cell Life Sci 1:1–5
Svedsen H, Beszczynska-Moller A, Hagen JO, Lefauconnier B, Tverberg V, Gerland S, Orbok JB, Bischof K, Papucci C, Zajaczkowski M, Azzolini R, Bruland O, Wiencke C, Winther JG, Dallmann W (2002) The physical environment of Kongsfjorden-Krossfjorden, an Arctic fjord system in Svalbard. Polar Res 21(1):133–166
Hop H, Pearson T, Hegseth EN, Kovacs KM, Wiencke C, Kwasniewski S, Eiane K, Mehlum F, Gulliksen B, Wlodarska-Kowalczuk M, Lydersen C, Weslawski JM, Cochrane S, Gabrielsen GW, Leakey RJG, Lønne OJ, Zajaczkowski M, Falk-Petersen S, Kendall M, Wängberg SÅ, Bischof K, Voronkov AY, Kovaltchouk NA, Wiktor J, Poltermann M, di Prisco G, Papucci C, Gerland S (2002) The marine ecosystem of Kongsfjorden, Svalbard. Polar Res 21(1):167–208
Martinez JL, Baquero F (2000) Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother 44(7):1771–1777
Xiong W, Sun Y, Zhang T, Ding X, Li Y, Wang M, Zeng Z (2015) Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China. Microb Ecol 70:425–432
Santos SS, Pardal S, Proença DN, Lopes RJ, Ramos JA, Mendes L, Morais PV (2012) Diversity of cloacal microbial community in migratory shorebirds that use the Tagus estuary as stopover habitat and their potential to harbor and disperse pathogenic microorganisms. FEMS Microbiol Ecol 82(1):63–74
Segawa T, Takeuchi N, Rivera A, Yamada A, Yoshimura Y, Barcaza G, Shinbori K, Motoyama H, Kohshima S, Ushida K (2013) Distribution of antibiotic resistance genes in glacier environments. Environ Microbiol Rep 5(1):127–134
Houndt T, Ochman H (2000) Long term shifts in patterns of antibiotic resistance in enteric bacteria. Appl Environ Microbiol 66:5406–5409
Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ (2016) The structure and diversity of human, animal and environmental resistomes. Microbiome 4:1–15
Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R (2018) Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci 115(15):E3463–E3470
Li B, Yang Y, Ma L, Ju F, Guo F, Tiedje JM, Zhang T (2015) Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J 9:2490–2502
Winker K, McCracken KG, Gibson DD, Pruett CL, Meier R, Huettmann F, Wege M, Kulikova IV, Zhuravlev YN, Perdue ML, Spackman E, Suarez DL, Swayne DE (2007) Movements of birds and avian influenza from Asia into Alaska. Emerg Infect Dis 13(4):547–552
Hernando-Amado S, Coque TM, Baquero F, Martínez JL (2019) Defining and combating antibiotic resistance from one health and Global Health perspectives. Nat Microbiol 4:1432–1442
Zhang Y, Gu AZ, He M, Li D, Chen J (2017) Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ Sci Technol 51(1):570–580
Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, Goorhuis A, Grobusch MP, Lashof AMO, Molhoek N, Schultsz C, Stobberingh EE, Verbrugh HA, de Jong MD, Melles DC, Penders J (2017) Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis 17(1):78–85
Tripathi V, Cytryn E (2017) Impact of anthropogenic activities on the dissemination of antibiotic resistance across ecological boundaries. Essays Biochem 61(1):11–21
Yuan L, Sun L, Long N, Xie Z, Wang Y, Liu X (2010) Seabirds colonized Ny-Ålesund, Svalbard, Arctic ~ 9,400 years ago. Polar Biol 33:683–691
Yang Z, Yuan L, Xie Z, Wang J, Li Z, Tu L, Sun L (2020) Historical records and contamination assessment of potential toxic elements (PTEs) over the past 100 years in Ny-Ålesund. Svalbard Environ Pollut 266:115205
Acknowledgements
This work was supported by the National Centre for Polar and Ocean Research, Ministry of Earth Sciences (MoES), Government of India, under the PACER-POP programme (NCPOR/2019/PACER-POP/BS-02). The authors are grateful to Director, National Centre for Polar and Ocean Research and Head, Department of Marine Biology, Microbiology & Biochemistry, Cochin University of Science and Technology, for providing the necessary facilities to carry out the work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Responsible Editor: Luiz Henrique Rosa.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
S, V., T, J., E, A.P. et al. Antibiotic resistance of heterotrophic bacteria from the sediments of adjoining high Arctic fjords, Svalbard. Braz J Microbiol (2024). https://doi.org/10.1007/s42770-024-01368-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42770-024-01368-0