Abstract
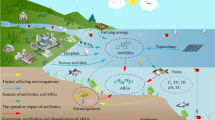
Environmental antibiotic resistance has drawn increasing attention due to its great threat to human health. In this study, we investigated concentrations of antibiotics (tetracyclines, sulfonamides and (fluoro)quinolones) and abundances of antibiotic resistance genes (ARGs), including tetracycline resistance genes, sulfonamide resistance genes, and plasmid-mediated quinolone resistance genes, and analyzed bacterial community composition in aquaculture environment in Guangdong, China. The concentrations of sulfametoxydiazine, sulfamethazine, sulfamethoxazole, oxytetracycline, chlorotetracycline, doxycycline, ciprofloxacin, norfloxacin, and enrofloxacin were as high as 446 μg kg−1 and 98.6 ng L−1 in sediment and water samples, respectively. The relative abundances (ARG copies/16S ribosomal RNA (rRNA) gene copies) of ARGs (sul1, sul2, sul3, tetM, tetO, tetW, tetS, tetQ, tetX, tetB/P, qepA, oqxA, oqxB, aac(6′)-Ib, and qnrS) were as high as 2.8 × 10−2. The dominant phyla were Proteobacteria, Bacteroidetes, and Firmicutes in sediment samples and Proteobacteria, Actinobacteria and Bacteroidetes in water samples. The genera associated with pathogens were also observed, such as Acinetobacter, Arcobacter, and Clostridium. This study comprehensively investigated antibiotics, ARGs, and bacterial community composition in aquaculture environment in China. The results indicated that fish ponds are reservoirs of ARGs and the presence of potential resistant and pathogen-associated taxonomic groups in fish ponds might imply the potential risk to human health.



Similar content being viewed by others
References
Wright GD (2010) Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol 13:589–594
Finley RL, Collignon P, Larsson DJ, McEwen SA, Li X-Z, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E (2013) The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis 57:704–710
D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R (2011) Antibiotic resistance is ancient. Nature 477:457–461
Zhu Y-G, Johnson TA, Su J-Q, Qiao M, Guo G-X, Stedtfeld RD, Hashsham SA, Tiedje JM (2013) Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A 110:3435–3440
Yang Y, Li B, Zou S, Fang HH, Zhang T (2014) Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res 62:97–106
Shah SQ, Cabello FC, L'Abée-Lund TM, Tomova A, Godfrey HP, Buschmann AH, Sørum H (2014) Antimicrobial resistance and antimicrobial resistance genes in marine bacteria from salmon aquaculture and non‐aquaculture sites. Environ Microbiol 16:1310–1320
Su JQ, Wei B, Xu CY, Qiao M, Zhu YG (2014) Functional metagenomic characterization of antibiotic resistance genes in agricultural soils from China. Environ Int 65:9–15
Amos G, Zhang L, Hawkey P, Gaze W, Wellington E (2014) Functional metagenomic analysis reveals rivers are a reservoir for diverse antibiotic resistance genes. Vet Microbiol 171:441–447
Shah SQ, Colquhoun DJ, Nikuli HL, Sørum H (2012) Prevalence of antibiotic resistance genes in the bacterial flora of integrated fish farming environments of Pakistan and Tanzania. Environ Sci Technol 46:8672–8679
Heuer OE, Kruse H, Grave K, Collignon P, Karunasagar I, Angulo FJ (2009) Human health consequences of use of antimicrobial agents in aquaculture. Clin Infect Dis 49:1248–1253
Seyfried EE, Newton RJ, Rubert KF IV, Pedersen JA, McMahon KD (2010) Occurrence of tetracycline resistance genes in aquaculture facilities with varying use of oxytetracycline. Microb Ecol 59:799–807
Sapkota A, Sapkota AR, Kucharski M, Burke J, McKenzie S, Walker P, Lawrence R (2008) Aquaculture practices and potential human health risks: current knowledge and future priorities. Environ Int 34:1215–1226
Dang STT, Petersen A, Van Truong D, Chu HTT, Dalsgaard A (2011) Impact of medicated feed on the development of antimicrobial resistance in bacteria at integrated pig-fish farms in Vietnam. Appl Environ Microbiol 77:4494–4498
Hoa PTP, Managaki S, Nakada N, Takada H, Shimizu A, Anh DH, Viet PH, Suzuki S (2011) Antibiotic contamination and occurrence of antibiotic-resistant bacteria in aquatic environments of northern Vietnam. Sci Total Environ 409:2894–2901
Di Cesare A, Luna GM, Vignaroli C, Pasquaroli S, Tota S, Paroncini P, Biavasco F (2013) Aquaculture can promote the presence and spread of antibiotic-resistant Enterococci in marine sediments. PLoS One 8:e62838
Gao P, Mao D, Luo Y, Wang L, Xu B, Xu L (2012) Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res 46:2355–2364
Li B, Zhang T, Xu ZY, Fang HHP (2009) Rapid analysis of 21 antibiotics of multiple classes in municipal wastewater using ultra performance liquid chromatography-tandem mass spectrometry. Anal Chim Acta 645:64–72
Pei R, Kim S-C, Carlson KH, Pruden A (2006) Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40:2427–2435
Aminov R, Garrigues-Jeanjean N, Mackie R (2001) Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol 67:22–32
Ng L-K, Martin I, Alfa M, Mulvey M (2001) Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes 15:209–215
Xia L-N, Li L, Wu C-M, Liu Y-Q, Tao X-Q, Dai L, Qi Y-H, Lu L-M, Shen J-Z (2010) A survey of plasmid-mediated fluoroquinolone resistance genes from Escherichia coli isolates and their dissemination in Shandong, China. Foodborne Pathog Dis 7:207–215
Kim HB, Wang M, Park CH, Kim E-C, Jacoby GA, Hooper DC (2009) oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother 53:3582–3584
Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC (2006) Prevalence in the United States of aac (6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955
Cattoir V, Poirel L, Rotimi V, Soussy C-J, Nordmann P (2007) Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 60:394–397
Bach H-J, Tomanova J, Schloter M, Munch J (2002) Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J Microbiol Methods 49:235–245
Xiong W, Sun Y, Ding X, Zhang Y, Zhong X, Liang W, Zeng Z (2015) Responses of plasmid-mediated quinolone resistance genes and bacterial taxa to (fluoro)quinolones-containing manure in arable soil. Chemosphere 119:473–478
Li S, Chou HH (2004) LUCY2: an interactive DNA sequence quality trimming and vector removal tool. Bioinformatics 20:2865–2866
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen A, McGarrell DM, Marsh T, Garrity GM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Takasu H, Suzuki S, Reungsang A, Viet PH (2011) Fluoroquinolone (FQ) contamination does not correlate with occurrence of FQ-resistant bacteria in aquatic environments of Vietnam and Thailand. Microbes Environ 26:135–143
Babić S, Periša M, Škorić I (2013) Photolytic degradation of norfloxacin, enrofloxacin and ciprofloxacin in various aqueous media. Chemosphere 91:1635–1642
Córdova-Kreylos AL, Scow KM (2007) Effects of ciprofloxacin on salt marsh sediment microbial communities. ISME J 1:585–595
Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI (2011) Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158
Tamminen M, Karkman A, Lõhmus A, Muziasari WI, Takasu H, Wada S, Suzuki S, Virta M (2010) Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ Sci Technol 45:386–391
Smalla K, Heuer H, Götz A, Niemeyer D, Krögerrecklenfort E, Tietze E (2000) Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl Environ Microbiol 66:4854–4862
Perreten V, Boerlin P (2003) A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother 47:1169–1172
Speer BS, Shoemaker NB, Salyers AA (1992) Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev 5:387–399
Roberts MC (2005) Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203
Agersø Y, Pedersen AG, Aarestrup FM (2006) Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J Antimicrob Chemother 57:832–839
Luo Y, Mao D, Rysz M, Zhou Q, Zhang H, Xu L, Alvarez JJ (2010) Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ Sci Technol 44:7220–7225
Graham DW, Olivares-Rieumont S, Knapp CW, Lima L, Werner D, Bowen E (2010) Antibiotic resistance gene abundances associated with waste discharges to the Almendares River near Havana, Cuba. Environ Sci Technol 45:418–424
Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegård B, Söderström H, Larsson DJ (2011) Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One 6:e17038
Marti E, Jofre J, Balcazar JL (2013) Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS One 8:e78906
Akinbowale OL, Peng H, Barton MD (2007) Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J Appl Microbiol 103:2016–2025
Doughari HJ, Ndakidemi PA, Human IS, Benade S (2011) The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ 26:101–112
Ho HT, Lipman LJ, Gaastra W (2006) Arcobacter, what is known and unknown about a potential foodborne zoonotic agent! Vet Microbiol 115:1–13
Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, Harmsen WS, Zinsmeister AR (2011) The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol 107:89–95
Boto L (2010) Horizontal gene transfer in evolution: facts and challenges. Proc R Soc B Biol Sci 277:819–827
Cabello FC, Godfrey HP, Tomova A, Ivanova L, Dölz H, Millanao A, Buschmann AH (2013) Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ Microbiol 15:1917–1942
Acknowledgments
This work was supported by grant 2013CB 127203 from the National Key Basic Research Program of China and grant U1031004 from the National Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Wenguang Xiong and Yongxue Sun contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Tables S1
(DOC 86 kb)
Rights and permissions
About this article
Cite this article
Xiong, W., Sun, Y., Zhang, T. et al. Antibiotics, Antibiotic Resistance Genes, and Bacterial Community Composition in Fresh Water Aquaculture Environment in China. Microb Ecol 70, 425–432 (2015). https://doi.org/10.1007/s00248-015-0583-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0583-x