Abstract
Wastewater-based epidemiology has been described as a valuable tool for monitoring the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a community. However, there is no consensus on the best concentration method to allow reliable detection of SARS-CoV-2 in this matrix, considering different laboratory facilities. This study compares two viral concentration methods, ultracentrifugation (ULT) and skimmed-milk flocculation (SMF), for detecting SARS-CoV-2 in wastewater samples. The analytical sensitivity (limits of detection and quantification [LoD/LoQ]) of both methods was evaluated using a bovine respiratory syncytial virus (BRSV) as a surrogate. Three different approaches were conducted to establish LoD of each method based on the assays on the standard curve (ALoDsc), on the dilution of internal control (ALoDiC), and the processing steps (PLoD). For PLoD, ULT method had the lowest value (1.86 × 103 genome copy/microliter [GC/µL]) when compared to the SMF method (1.26 × 107 GC/µL). The LoQ determination showed a mean value of 1.55 × 105 GC/µL and 3.56 × 108 GC/µL to ULT and SMF, respectively. The detection of SARSCoV-2 in naturally contaminated wastewater revealed 100% (12/12) and 25% (3/12) of detection using ULT and SMF with quantification ranging from 5.2 to 7.2 log10 genome copy/liter (GC/L) and 5.06 to 5.46 log10 GC/L, respectively. The detection success rate of BRSV used as an internal control process was 100% (12/12) for ULT and 67% (8/12) for SMF, with an efficiency recovery rate ranging from 12 to 38% and 0.1 to 5%, respectively. Our data consolidates the importance of assessing the methods used; however, further analysis should be carried out to improve low-cost concentration methodologies, essential for use in low-income and developing countries.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In January 2020, the world was surprised by the World Health Organization’s (WHO) declaration of an outbreak of a severe acute respiratory syndrome caused by a new beta-coronavirus, coronavirus-2 (SARS-CoV-2), thus announcing the beginning of the global pandemic of coronavirus disease 2019 (COVID-19). Although the transmission of SARS-CoV-2 is mainly by respiratory droplets [1, 2], it is well known that SARS-CoV-2-infected patients shed viral particles in feces before, during, and after COVID-19 symptoms for a few days to several weeks [3,4,5] Based on this knowledge, studies have demonstrated the utility of wastewater-based epidemiology (WBE) as a complementary tool to massive community testing in controlling COVID-19 [6]. This approach has been used since the beginning of the pandemic when studies were performed worldwide providing environmental data to early warning of an increase in virus transmission in a community [7,8,9,10,11,12,13,14,15,16,17].
Despite the diversity of available protocols performed in those studies, there is still no consensus on the best methodology used to concentrate and recover SARS-CoV-2 from wastewater samples, mainly due to the high complexity of the matrix associated with analytical sensitivity that involves a sequence of steps that can directly interfere with viral detection and quantification [18]. In 2021, the Pan American Health Organization published a technical guide recommending concentration methods for Latin American countries, such as ultracentrifugation, adsorption and precipitation with aluminum hydroxide, flocculation with skimmed milk, ultrafiltration, and adsorption on charged membrane followed by ultrafiltration. The guide also recommended the use of a surrogate virus to evaluate the concentration method by calculating the recovery efficiency, in addition to the fact that all technical procedures are performed in a biosafety level 2 (NB2) laboratory [19].
Ultracentrifugation (ULT) method has been used by many laboratories to concentrate and recover enteric viruses from these matrices [8, 16, 20,21,22]. The method is a straightforward factor limiting the high cost of an ultracentrifuge, especially for research laboratories in low- and middle-income countries. As an alternative, a low-cost method based on skimmed milk flocculation (SMF), previously described for recovering viruses from seawater, has been applied for different environmental matrices as sewage [15, 23,24,25,26,27,28]. However, detecting enveloped viruses, such as SARS-CoV-2, which can be more easily degraded in water environments, is an issue to observe for both methods [29], as well as, the physical–chemical characteristics of wastewater that may vary according to the habits and size of the population served and thus interfering with viral detection methods [15, 29]. To avoid all these difficulties, using an internal control process, as the target virus, is beneficial to ensure the efficiency of all stages of the process. For SARS-CoV-2, bovine respiratory syncytial virus (BRSV) has been used since its genome consists of a single-stranded RNA molecule, with an envelope in its structure [16, 18].
This study aimed to compare ULT and SMF concentration methods for recovery of SARS-CoV-2 from wastewater samples determining the limits of detection (LoD), as well as the limit of quantification (LoQ) and the efficient recovery of both methods using BRSV as a surrogate virus [8, 18, 30,31,32]. In order to establish the LoD for each method, three approaches were performed based on the standard curve (ALoDsc), on the dilution of the internal control (ALoDiC), and on the processing steps (PLoD) [18, 30]. After all, detection of the natural contamination by SARS-CoV-2 was performed using both concentration methods.
Materials and methods
Study area and sampling collection points
In January 2021, samples of raw sewage were collected in the sanitary sewage system in the city of Niteroi, state of Rio de Janeiro, located in the southeastern region of Brazil. The city has around 515,000 inhabitants and based on data from the National Sanitation Information System, 94.5% of the population is attended by the sewage collection and treatment service [33].
Sewage sample collection was obtained at eight different wastewater treatment plants (WWTP) and four sewer pipes (n = 12), being carried out in a compound way in 10 h, with a total volume of one liter per sample. The samples were placed in sterile polypropylene bottles and kept at 4 °C for 24 h until processing. The collection was carried out during the highest effluent flow in each sample point.
For spiking experiments, a pool of 3 L of sewage containing 250 mL of each collection point was prepared for LoD/LoQ triplicate assays. All 12 samples were investigated for SARS-CoV-2 occurrence in those matrices.
Virus surrogate
BRSV inoculum was prepared from the lyophilized Inforce 3 intranasal cattle vaccine (Zoetis-Parsippany, NJ, USA), following the manufacturer’s recommendation. The resuspended inoculum was quantified, and aliquots of 1 mL containing 107 genome copy/microliter (GC/µL) were stored at − 20 °C until use. BRSV inoculum was quantified at each use by RT-qPCR assay.
Viral concentration
Before the concentration step, each sample was inoculated with BRSV (500 µL), homogenized, and two aliquots with volumes of 42 and 50 mL were concentrated by ULT and SMF, respectively.
As previously described, the ULT method was performed [20]. Briefly, 42 mL of wastewater samples were centrifuged at 100,000 × g for 60 min at 4 °C. After discarding the supernatant, the pellet was dissolved in 4 mL of 0.25 N glycine buffer (pH 9.5) and incubated at 4 °C for 30 min, under stirring. Then, 4 mL of 2 × phosphate-buffered saline (PBS, pH 7.2) added and then clarified by centrifugation at 12,000 × g for 20 min. Finally, supernatant samples (~ 8 mL) were centrifuged at 100,000 × g for 60 min at 4 °C, and viral particles were suspended in 400 μL of 1 × PBS (pH 7.2).
The SMF was performed as previously described [24]. Briefly, 100 mL of glycine buffer 0.25 N, pH 9.5 (1:9, v/v) was added to 50 mL of the sewage sample and stirred for 30 min in ice, and centrifuged at 8000 × g for 30 min at 4 °C. The supernatant (150 mL) was transferred to a new bottle, and the pH was adjusted to 3.5 with HCl 6 N. The pre-flocculated skimmed-milk solution (1%, w/v) was added to the supernatant to obtain a final concentration of 0.01% (w/v). Samples were stirred for 8 h at room temperature, and flakes were pelleted by centrifugation at 8000 × g for 30 min at 4 °C. The supernatant was carefully removed, and the pellet was dissolved in 1000 μL of phosphate buffer (pH 7.5). Final concentrated sample volume was immediately stored at − 80 °C until viral nucleic acid extraction.
Nucleic acid extraction
Viral nucleic acid was extracted from 140 μL of each sample using the QIAamp® Viral RNA Mini kit (QIAGEN, CA, USA) in a QIAcube® automated system (QIAGEN), as indicated by the manufacturer, obtaining a final volume of 60 μL. The extracted samples were immediately stored at − 80 °C for later use.
Viral detection and quantification
Primers and probes used for BRSV and SARS-CoV-2 quantification were based on the TaqMan® protocol. BRSV detection was performed by targeting the conserved region of the N gene, according to the protocol previously described [34]. The reactions were performed using 5 µL of RNA extracted, 10 µL of the AgPath-ID™ One-Step RT-PCR Reagents kit, 0.8 µL primers BRSV-F and BRSV-R (final concentration 0.4 µM), and probe BRSV-P (final concentration 0.2 µM). The detection of SARS-CoV-2 was performed by targeting the N2 region (nucleocapsid gene) [35]. It was used 5 μL of RNA, 10 µL of SuperScriptTM III Platinum TM kit qRT-qPCR One-Step (Invitrogen), and 1.5 µL of primer and probe mix for the N2 region (2019-nCoV RUO Kit, Integrated DNA Technologies, Coralville, IA, USA). Reactions were performed in the ABI PRISM 7500 (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions. The reaction cycle was programmed for 30 min at 50 °C, 10 min at 95 °C, and 40 cycles of 15 s at 95 °C, 30 s at 60 °C.
The quantification of BRSV and SARS-CoV-2 RNA was estimated using a standard curve, in which serial 10-fold dilutions were used, with copies ranging from 100 to 106 GC/µL and 101 to 105 GC/µL, respectively, of a fragment of double-stranded DNA (gBlock Gene Fragment, Integrated DNA Technologies) containing the sequence of the specific target amplification region. Samples were considered positive when at least two of the four wells (diluted and undiluted) had a cycle threshold (Ct) value lower than 40.
To preserve samples from cross-contamination, quality control of molecular procedures was ensured with the use of different rooms in each of the processing activities. Negative process controls (RNAse-free water) and no template controls were included in each RT-qPCR running.
LoD/LoQ determination
The LoD was defined as the smallest number of genomic copies (GC) of a given target capable of generating a positive result in the RT-qPCR reaction in at least 95% replicates while LoQ as the smallest amount of detection and quantification with precision and accuracy, with pre-defined variability and coefficient of variation (CV) < 25% [30, 36].
To explore all the data generated, we conducted three different approaches to estimate the LoD: (i) assay determined by the standard curve (ALoDsc), (ii) assay by the serial dilution of the internal control (ALoDic), and (iii) assay based on entire sampling process (PLoD) [18, 32].
The ALoDsc was determined based on the 10-fold serial dilution of the standard curve used (1.6 × 102 to 106 GC/µL). To estimate the ALoDic, a 10-fold serial dilution of the internal control was applied: 100 to 107 GC/µL. The estimated values for PLoD and LoQ were obtained by evaluating the results performed in triplicate (A, B, and C). For each replicate, eight aliquots were spiked with a 500-µL serial 10-fold dilution of the BRSV inoculum (with theoretical concentrations ranging from 100 to 107 GC/µL.). After the extraction step, the viral RNA was subjected to detection by the RT-qPCR assay, evaluating undiluted and diluted replicas. ALoDsc, ALoDic, and PLoD were estimated using a logistic regression model (equation p). It was possible to identify the concentration at which the probability of a positive response was at least 95%.
The LoQ was determined to follow a previously proposed model [37]. The probability distribution of the log-normal type was considered for the generated data, where the coefficient of variation (CV) (geometric) was based on the standard deviation (SD) of the concentrations on a logarithmic basis, according to equation CV.
where p is probability of positive responses; L is proportionality constant; e is Euler’s number; K is model inflection rate; (log GC/µL) is concentration (number of genomic copies/ microliter); SD standard deviation; Ln (CG/µL) is natural logarithm of concentrations.
Internal process control and recovery efficiency
To evaluate the efficiency of the viral concentration methods, samples were seeded with a 500-µL of BRSV vaccine suspension containing 107 GC/µL as an internal control process.
The recovery efficiency was calculated considering the values recovered in the RT-qPCR assays (raw data). Then, the cutoff values obtained in ALoDsc, ALoDic, and PLoD were applied.
The recovery efficiency (%) was calculated according to equation E.
Statistical analysis
Mathematical models to obtain ALoD, PLoD, and LoQ values were performed using the Microsoft Excel software (2013). Descriptive statistical analysis, Mann Whitney test, and elaboration of figures were performed using the GraphPad Prism v 9.3.0 software (GraphPad Software, San Diego, CA, USA).
Statistical analyzes were performed using a significance level of 0.05 with 12 samples for the study.
Results
LoD/LoQ determination
Initially, we searched for the natural occurrence of BRSV in the wastewater samples demonstrating virus’s absence in the environmental samples collected. Following, the analytical sensitivity of the RT-qPCR assay was assessed by the values established for ALoDsc and ALoDic with both results presenting the same order of magnitude (102 GC/µL) with average values of 6.6 × 102 GC/µL and 1.7 × 102 GC/µL, respectively. The results obtained for PLoD revealed ULT with average values four logs lower when compared to the SMF method. The ULT LoQ determination showed an average of 1.86 × 103 GC/µL, while SMF results showed a mean value of 3.56 × 108 GC/µL (Table 1).
Internal process control recovery
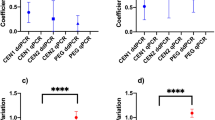
BRSV were detected in 100% (12/12) of the samples when concentrated by ULT and 92% (11/12) by the SMF method. The recovery rates ranged from 12 to 38%, mean 19%, for ULT, and 0.1 to 5%, 2%, for SMF. The samples concentrated by ULT showed viral concentrations ranging from 7.4 to 7.9 log10 genome copy/microliter (GC/µL) ± 0.17. On the other hand, the viral quantification of the samples concentrated by SMF showed lower values ranging from 0.9 to 6.6 log10 GC/µL ± 1.6 (Fig. 1A).
Once the LoD values were established, we calculated the recovery efficiency by applying the limits obtained by ALoDsc, ALoDic, and PLoD (Fig. 1B, C, and D). After applying the cutoff values, the ULT concentration method showed the same detection success (100%) in all comparative evaluations. However, significant differences were observed in the recovery efficiency of samples concentrated by SMF, varying between 17 and 33% after the cut made for the limits obtained by ALoDsc and ALoDic, respectively. None of the samples concentrated by SMF showed values higher than the established limit for PLoD. The discrepancy of the results obtained between the concentration methods was statistically confirmed using the Mann–Whitney test, and the statistical significance was determined by the p-value, where p < 0.0001 in the raw results; p = 0.0011 obtained after cutting by ALoDsc; p = 0.0220 for ALoDsc.
As for the evaluation of quantification, 100% (12/12) of the samples concentrated by ULT showed values higher than the LoQ, while none of the samples concentrated by SMF showed values greater than or equal to the LoQ.
Detection and quantification of SARS-CoV-2
SARS-CoV-2 was detected in 100% (12/12) of samples concentrated by ULT with viral load ranging from 5.2 to 7.2 log10 GC/L ± 0.6. Using the SMF, only three of the twelve samples (25%) were positive for SARS-CoV-2, and the viral concentration was 5.06, 5.08, and 5.46 log10 GC/L ± 0.2. Samples detected by SMF do not correspond to the samples that presented the highest quantification values when concentrated by ULT (6.54, 7.22, and 7.15 log10 GC/L, respectively) (Fig. 2).
Discussion
A comparative study comparing the analytical sensitivity (LoD/LoQ) between two viral concentration methods revealed the importance of quantification of those limits using different approaches (ALoDsc, ALoDic, and PLoD) for assessing the recovery efficiency of a specific virus in each matrix such as wastewater. Here, we demonstrated that ULT presented a better analytical sensitivity and recovery efficiency compared to SMF for BRSV, as well as for detecting SARS-CoV-2 from naturally contaminated samples.
To estimate the detection of BRSV RNA, whose probability of a positive response must be ≥ 95% [36, 38, 39], we used qPCR data as a categorical variable allowing the adjustment of the sigmoid function given a linear combination of different attributes. Our results regarding ALoD were lower than obtained in a previous study (4.42 GC/reaction) where a multiple viral concentration protocols for SARS-CoV-2 in wastewater samples was used [18]. However, different studies conducted to evaluate concentration methods using other surrogate viruses presented values for ALoD ranging from 5 × 103 to 6.6 × 103 GC/reaction [8, 15, 26, 30, 32].
Since ALoD approach does not evaluate all the sample processing steps, not allowing the comparison between the concentration methods, we added PLoD concept to this analysis. PLoD results demonstrated that ULT method detected values lower (103 GC/µL) than those obtained for ALoDsc and ALoDic, showing that the methodology used is able to remove enzyme reaction inhibitors present in this matrix. In contrast, SMF PLoD was considerably higher (107 GC/µL) revealing a lower capacity to minimize the inhibitory or the effect of employing milk protein to concentrate virus. Using the adsorption extraction (AE) concentration method, Ahmed et al. [32] obtained values of PLoD using RT-qPCR and RT-dPCR for SARS-CoV-2 ranging from 3.9 to 6.6 GC/50 mL, respectively.
Concerning LoQ analysis, we also demonstrated that ULT was able to detect viral RNA at lower concentrations when compared to SMF using CV of 25%. Previous studies using CV of 35% have established values for LoQ for different concentration methods and surrogate virus, obtaining values from 4.56 × 104 to 1.22 × 105 GC/reaction [26]; 2.56 × 103 to 2.56 × 105 GC/reaction [31]; 2.35 × 104 GC/L to 3.56 × 103 GC/L [30]; 1,000 and 100 gene copies [15]; 5 GC/reaction [8].
Using estimated analytical sensitivity, ULT maintained the same results, for both detecting success rate and recovery efficiency. On the other hand, samples concentrated by SMF had a stronger influence on the results. Similar results were previous reported using ULT and BRSV, but our results were higher when compared using heat-inactivated SARS-CoV-2 as surrogate [16, 40].
Despite the low recovery efficiency of SFM using BRSV (1% ± 1.7), our results were similar previous studies evaluating this method with different surrogate viruses. For instance, Salvo et al. [28] found a recovery rate of 1.9% and 8.9% for BCoV for undiluted and diluted samples, respectively; Monteiro et al. [26] found a recovery efficiency of 14% for PEDV; and Barril et al. [23] found a recovery rate of 1% for feline coronavirus. In another study, using human coronavirus as a surrogate, it was demonstrated a slightly improvement in the recovery efficiency of SMF method, with results ranging from 9 to 4% using Vertrel™ treatment, and 6 to 2.6% without treatment [15, 27]. The SMF method has been described as a low cost method being used to detect different enteric viruses in several aquatic matrices with satisfactory results without the use of Vertrel™ treatment [41,42,43]. Although with initial high cost relating to the ultracentrifuge acquisition, the ULT method proved to be efficient to detect SARS-CoV-2 [16] (Prado et al., 2021). More recently, Zheng et al. [40] adapted the ULT method using only one ultracentrifugation step achieving an average recovery efficiency of 25.4% using inactivated SARS-CoV-2.
In a recent study comparing different protocols for SARS-CoV-2 detection, the method of Centrifuge-UF-15 (100 kDa) presented a better performance with 57% (12/21) [18]. The variability in the detection of SARS-CoV-2 RNA between different methods was associated with the fact that the viruses were found at levels close to the LoD of the evaluated techniques, mainly due to effluent dilution, low persistence of the virus in the environment, and/or high presence of inhibitory charge [15].
Our study comparing those previously standardized viral concentration protocols for non-enveloped enteric viruses [20, 25] aimed to evaluate their performance in detecting and quantifying enveloped viruses so a single protocol could be used in environmental surveillance studies using wastewater as a research matrix. ULT overcome SMF method, presenting results in concordance with recent published data [8, 16, 40]. The attempt to optimize the SMF method, previously carried out by Philo [27], showed a small increase in the recovery rate, suggesting that further studies aimed at reducing inhibitors may allow the use of this method.
Conclusion
Based on our analysis, the ULT method showed higher analytical sensitivity for the detection of enveloped viruses compared to samples concentrated by SMF, being this result corroborated by the high percentage of SARS-CoV-2-positive samples detected in naturally contaminated samples than using the SMF method.
References
Chia PY, Coleman KK, Tan YK, Ong SWX, Gum M, Lau SK, Lim XF, Lim AS, Sutjipto S, Lee PH et al (2020) Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun 11:2800. https://doi.org/10.1038/s41467-020-16670-2
Liu Y, Ning Z, Chen Y, Guo M, Liu Y, Gali NK, Sun L, Duan Y, Cai J, Westerdahl D et al (2020) Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 582:557–560. https://doi.org/10.1038/s41586-020-2271-3
Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, Yang Y, Liu B, Wang W, Wei C et al (2020) The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol 92:833–840. https://doi.org/10.1002/jmv.25825
Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TWH, Tam AR et al (2020) Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology 159:81–95. https://doi.org/10.1053/j.gastro.2020.03.065
Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J et al (2020) Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 26:502–505. https://doi.org/10.1038/s41591-020-0817-4
Smyth DS, Trujillo M, Gregory DA, Cheung K, Gao A, Graham M, Guan Y, Guldenpfennig C, Hoxie I, Kannoly S et al (2022) Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat Commun 13:635. https://doi.org/10.1038/s41467-022-28246-3
Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, Choi PM, Kitajima M, Simpson SL, Li J et al (2020) First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 728:138764. https://doi.org/10.1016/j.scitotenv.2020.138764
Ahmed W, Bertsch PM, Bivins A, Bibby K, Farkas K, Gathercole A, Haramoto E, Gyawali P, Korajkic A, McMinn BR et al (2020) Comparison of virus concentration methods for the RT-QPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci Total Environ 739:139960. https://doi.org/10.1016/j.scitotenv.2020.139960
Daughton CG (2020) Wastewater surveillance for population-wide Covid-19: the present and future. Sci Total Environ 736:139631. https://doi.org/10.1016/j.scitotenv.2020.139631
Farkas K, Hillary LS, Malham SK, McDonald JE, Jones DL (2020) Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr Opin Environ Sci Health 17:14–20. https://doi.org/10.1016/j.coesh.2020.06.001
Hart OE, Halden RU (2020) Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci Total Environ 730:138875. https://doi.org/10.1016/j.scitotenv.2020.138875
Mao K, Zhang K, Du W, Ali W, Feng X, Zhang H (2020) The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr Opin Environ Sci Health 17:1–7. https://doi.org/10.1016/j.coesh.2020.04.006
Medema G, Been F, Heijnen L, Petterson S (2020) Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr Opin Environ Sci Health 17:49–71. https://doi.org/10.1016/j.coesh.2020.09.006
Nghiem LD, Morgan B, Donner E, Short MD (2020) The COVID-19 pandemic: considerations for the waste and wastewater services sector. Case Stud Chem Environ Eng 1:100006. https://doi.org/10.1016/j.cscee.2020.100006
Philo SE, Keim EK, Swanstrom R, Ong AQW, Burnor EA, Kossik AL, Harrison JC, Demeke BA, Zhou NA, Beck NK et al (2021) A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci Total Environ 760:144215. https://doi.org/10.1016/j.scitotenv.2020.144215
Prado T, Fumian TM, Mannarino CF, Resende PC, Motta FC, Eppinghaus ALF, Vale VHC, Braz RMS, de Andrade JDSR, Maranhão AG et al (2021) Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res 191:116810. https://doi.org/10.1016/j.watres.2021.116810
Thompson JR, Nancharaiah YV, Gu X, Lee WL, Rajal VB, Haines MB, Girones R, Ng LC, Alm EJ, Wuertz S (2020) Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res 184:116181. https://doi.org/10.1016/j.watres.2020.116181
Kaya D, Niemeier D, Ahmed W, Kjellerup BV (2022) Evaluation of multiple analytical methods for SARS-CoV-2 surveillance in wastewater samples. Sci Total Environ 808:152033. https://doi.org/10.1016/j.scitotenv.2021.152033
PAHO - Pan American Health Organization (2021) Guía para el análisis y la cuantificación del SARS-CoV-2 en aguas residuales. OPS/CDE/CE/COVID-19/21-0014. PAHO, Washington, DC. https://iris.paho.org/bitstream/handle/10665.2/54698/CDECECOVID-19210014_spa.pdf?sequence=1&isAllowed=y
Pina S, Puig M, Lucena F, Jofre J, Girones R (1998) Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl Environ Microbiol 64:3376–3382. https://doi.org/10.1128/AEM.64.9.3376-3382.1998
Fumian TM, Leite JPG, Castello AA, Gaggero A, de Caillou MSL, Miagostovich MP (2010) Detection of rotavirus A in sewage samples using multiplex QPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J Virol Methods 170:42–46. https://doi.org/10.1016/j.jviromet.2010.08.017
Ye Y, Ellenberg RM, Graham KE, Wigginton KR (2016) Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ Sci Technol 50:5077–5085. https://doi.org/10.1021/acs.est.6b00876
Barril PA, Pianciola LA, Mazzeo M, Ousset MJ, Jaureguiberry MV, Alessandrello M, Sánchez G, Oteiza JM (2021) Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Sci Total Environ 756:144105. https://doi.org/10.1016/j.scitotenv.2020.144105
Calgua B, Rodriguez-Manzano J, Hundesa A, Suñen E, Calvo M, Bofill-Mas S, Girones R (2013) New methods for the concentration of viruses from urban sewage using quantitative PCR. J Virol Methods 187:215–221. https://doi.org/10.1016/j.jviromet.2012.10.012
Calgua B, Mengewein A, Grunert A, Bofill-Mas S, Clemente-Casares P, Hundesa A, Wyn-Jones AP, López-Pila JM, Girones R (2008) Development and application of a one-step low cost procedure to concentrate viruses from seawater samples. J Virol Methods 153:79–83. https://doi.org/10.1016/j.jviromet.2008.08.003
Monteiro S, Rente D, Cunha MV, Marques TA, Cardoso E, Álvaro P, Vilaça J, Ribeiro J, Silva M, Coelho N et al (2022) Recovery of SARS-CoV-2 from large volumes of raw wastewater is enhanced with the Inuvai R180 system. J Environ Manag 304:114296. https://doi.org/10.1016/j.jenvman.2021.114296
Philo SE, Ong AQW, Keim EK, Swanstrom R, Kossik AL, Zhou NA, Beck NK, Meschke JS (2022) Development and validation of the skimmed milk pellet extraction protocol for SARS-CoV-2 wastewater surveillance. Food Environ Virol. https://doi.org/10.1007/s12560-022-09512-5
Salvo M, Moller A, Alvareda E, Gamazo P, Colina R, Victoria M (2021) Evaluation of low-cost viral concentration methods in wastewaters: implications for SARS-CoV-2 pandemic surveillances. J Virol Methods 297:114249. https://doi.org/10.1016/j.jviromet.2021.114249
La Rosa G, Bonadonna L, Lucentini L, Kenmoe S, Suffredini E (2020) Coronavirus in water environments: occurrence, persistence and concentration methods — a scoping review. Water Res 179:115899. https://doi.org/10.1016/j.watres.2020.115899
Forés E, Bofill-Mas S, Itarte M, Martínez-Puchol S, Hundesa A, Calvo M, Borrego CM, Corominas LL, Girones R, Rusiñol M (2021) Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci Total Environ 768:144786. https://doi.org/10.1016/j.scitotenv.2020.144786
LaTurner ZW, Zong DM, Kalvapalle P, Gamas KR, Terwilliger A, Crosby T, Ali P, Avadhanula V, Santos HH, Weesner K et al (2021) Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res 197:117043. https://doi.org/10.1016/j.watres.2021.117043
Ahmed W, Bivins A, Metcalfe S, Smith WJM, Verbyla ME, Symonds EM, Simpson SL (2022) Evaluation of process limit of detection and quantification variation of SARS-CoV-2 RT-QPCR and RT-DPCR assays for wastewater surveillance. Water Res 213:118132. https://doi.org/10.1016/j.watres.2022.118132
SNIS–Sistema Nacional de Informações em Saneamento (2020) Série Histórica. Available: http://app4.mdr.gov.br/serieHistorica/. Accessed 4 Aug 2022
Boxus M, Letellier C, Kerkhofs P (2005) Real time RT-PCR for the detection and quantitation of bovine respiratory syncytial virus. J Virol Methods 125:125–130. https://doi.org/10.1016/j.jviromet.2005.01.008
Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, Lynch B, Malapati L, Burke SA, Harcourt J et al (2020) US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1654–1665. https://doi.org/10.3201/eid2608.201246
Burns M, Valdivia H (2008) Modelling the limit of detection in real-time quantitative PCR. Eur Food Res Technol 226:1513–1524. https://doi.org/10.1007/s00217-007-0683-z
Forootan A, Sjöback R, Björkman J, Sjögreen B, Linz L, Kubista M (2017) Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (QPCR). Biomolecular Detect Quantif 12:1–6. https://doi.org/10.1016/j.bdq.2017.04.001
Pavšič J, Žel J, Milavec M (2016) Assessment of the real-time PCR and different digital PCR platforms for DNA quantification. Anal Bioanal Chem 408:107–121. https://doi.org/10.1007/s00216-015-9107-2
Ricchi M, Savi R, Bolzoni L, Pongolini S, Grant IR, De Cicco C, Cerutti G, Cammi G, Garbarino CA, Arrigoni N (2016) Estimation of Mycobacterium avium subsp. paratuberculosis load in raw bulk tank milk in Emilia-Romagna region (Italy) by qPCR. Microbiol Open 5:551–559. https://doi.org/10.1002/mbo3.350
Zheng X, Deng Y, Xu X, Li S, Zhang Y, Ding J, On HY, Lai JCC, In Yau C, Chin AWH et al (2022) Comparison of virus concentration methods and RNA extraction methods for SARS-CoV-2 wastewater surveillance. Sci Total Environ 824:153687. https://doi.org/10.1016/j.scitotenv.2022.153687
Fioretti JM, Fumian TM, Rocha MS, Santos IAL, Carvalho-Costa FA, de Assis MR, Rodrigues JS, Leite JPG, Miagostovich MP (2018) Surveillance of noroviruses in Rio De Janeiro, Brazil: occurrence of new GIV genotype in clinical and wastewater samples. Food Environ Virol 10:1–6. https://doi.org/10.1007/s12560-017-9308-2
Macena LGP, Feitosa RC, Vieira CB, Araújo IT, Taniuchi M, Miagostovich MP (2021) Microbiological assessment of an urban lagoon system in the coastal zone of Rio de Janeiro, Brazil. Environ Sci Pollut Res 28:1170–1180. https://doi.org/10.1007/s11356-020-10479-8
Shubo T, Maranhão A, Ferreira FC, Fumian TM, Pimenta MMA, Morgado CRV, Toze S, Ahmed W, Sidhu J, Miagostovich MP (2021) Virological characterization of roof-harvested rainwater of densely urbanized low-income region. Food Environ Virol 13:412–420. https://doi.org/10.1007/s12560-021-09484-y
Acknowledgements
The authors would like to thank the Municipality of Niterói for supporting this research and the company Águas de Niterói for their support in collecting samples. They also thank the technical staff Sérgio de Silva e Mouta Júnior and Márcia Maria Araújo Pimenta (Oswaldo Cruz Foundation, RJ, Brazil) for helping with sample processing.
Funding
This work was supported by the Fundação de Amparo à Pesquisa do Rio de Janeiro (Faperj-E-26/211.311/2021), Conselho de Desenvolvimento Científico e Tecnológico (CNPq 306655/2018–7), and Instituto Oswaldo Cruz (PAEF3). This research study is under the scope of the activities of the Oswaldo Cruz Foundation (FIOCRUZ) as a Collaborating Centre of PAHO/WHO of Public and Environmental Health. SISGEN A2CBA1.
Author information
Authors and Affiliations
Contributions
Conceptualization: André Ribeiro, Tatiana Prado, Fernando Ferreira, and Marize Miagostovich; data curation: André Ribeiro; Formal analysis, André Ribeiro, and Eduardo Castro; funding acquisition: Camille Mannarino and Marize Miagostovich; investigation: André Ribeiro; Methodology, André Ribeiro, Tatiana Prado, Fernando Ferreira, Tulio Fumian, and Marize Miagostovich; resources: Camille Mannarino and Marize Miagostovich; software: Eduardo Castro; supervision: Marize Miagostovich; Validation, Eduardo Castro, and Marize Miagostovich; visualization: Camille Mannarino; writing — original draft: André Ribeiro; writing — review and editing: Camille Mannarino, Eduardo Castro, Tatiana Prado, Tulio Fumian, and Marize Miagostovich.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Jônatas Abrahão
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ribeiro, A.V.C., Mannarino, C.F., de Castro, E.S.G. et al. Assessment of virus concentration methods for detecting SARS-CoV-2 IN wastewater. Braz J Microbiol 54, 965–973 (2023). https://doi.org/10.1007/s42770-023-00941-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-00941-3