Abstract
The number of SARS-CoV-2 detection tests requested to the laboratories has dramatically increased together with an urgent need to release reliable responses in a very short time. The two options taken into consideration and analyzed in the current study were the point-of-care test (POCT) based on the nucleic acid amplification test (NAAT) and the Antigen (Ag) rapid test. The POCT-NAAT-based assay was compared with a rapid antigen test of nasopharyngeal swab samples. If the specimen tested positive, it was followed by viral load quantification and by the functional assessment of the residual infectivity. When the initial cycle threshold (Ct) was below 20 (100%), and in the range of 20–25 (92%) and of 25–30 (88%), a great concordance between the POCT-NAAT and the Ag test was observed. Moreover, the positivity of the antigen test was well correlated to a successful infection in vitro (78%), with greater concordance when the initial Ct below 20 or above 35 (100%) and in the range 20–25 (83%). Our findings showed that most of the swabs which tested positive using the antigen test were able to infect the cells in vitro, suggesting that probably only these samples hold residual infectivity and therefore an increased risk of virus transmission at the moment of being tested.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The nucleic acid amplification test (NAAT) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA mainly exploiting the reverse transcription-quantitative polymerase reaction (RT-qPCR) is the gold standard for the virus detection in clinical specimens and it is commonly conducted on nasopharyngeal swabs. The increased number of SARS-CoV-2 detection tests requested to the laboratories has dramatically raised the workload of the diagnostic procedures multiplying their efforts for the release of reliable responses in a very short time [1]. This need is crucial in the hospitals for the emergency room admission, hospitalization, or urgent surgery in order to avoid nosocomial outbreaks in the hospital wards [2]. Therefore, different rapid diagnostic tests have been developed and are actually present in the market. The point of care test (POCT) based on the RT-PCR platform is a valid option to efficiently obtain results in less than 1 h (e.g., GeneXpert Xpress SARS-CoV-2, Cepheid); meanwhile, exploiting the isothermal amplification procedure the data can be available in less than 30 min (e.g., ID NOw COVID-19 Abbott Diagnostics) so greatly accelerating the diagnostic process [3,4,5]. However, the main limitation of this procedure is related to the constrain of employing instruments just dedicated to one typology of kit and consumables provided by the same company. Moreover, since it is possible to run a few tests at a time, the POCT presents low processivity, thus representing a main limitation for large-scale use [5]. Antigen rapid test is another cheaper alternative to generate results in 10–30 min; nevertheless, this type of test presents a lower performance in terms of sensitivity [2]. Indeed, the medical agencies worldwide, European Centre for Disease Prevention and Control (ECDC), Centers for Disease Control and Prevention (CDC), and World Health Organization (WHO), recommended the use of rapid diagnostic tests [6,7,8] but requiring high performance, for example, ECDC indicates the use of tests showing ≥ 90% sensitivity and ≥ 97% specificity. Moreover, it is important to consider that this type of tests present high sensitivity in patients with high viral load (RT-PCR cycle threshold < 25) that primarily occurs in pre-symptomatic and early symptomatic phases (up to 5 days from symptoms onset) [8].
Although these types of patients are the leading cause of SARS-CoV-2 transmission to the neighbors, it should not be forgotten that also individuals with low viral load can be a source of infectious virions [9].
The main target of the antigen test, as well as of the RT-PCR-based assays, is the nucleocapsid (N gene) of SARS-CoV-2 since it is the most abundant and conserved protein in coronavirus [10].
The aim of the present study was the comparison of a POCT-NAAT-based assay with a rapid antigen test followed by the viral load quantification and by the functional assessment of the residual infectivity of the nasopharyngeal swab samples analyzed.
Materials and methods
Workflow of the experimental settings
Clinical nasopharyngeal swabs which resulted positive for the SARS-CoV-2 ORF1ab and N genes at the novel coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech Inc., Changsha, China) on the iPonatic Portable Molecule Workstation (AB Analitica, Padova, Italy) were tested for the viral antigen nucleocapsid, with the STANDARD F COVID-19 Ag FIA, immunofluorescence immunoassay on the Standard FX 2400 analyzer (SD BIOSENSOR, Suwon, Korea).
Then, the same samples were further quantified with a homemade RT-qPCR targeting the N gene with CDC specific primers and probe [11] on a 7500 Real-Time PCR Applied Biosystem platform (Thermo Fisher Scientific, Waltham, MA, USA); then, the residual infectivity of the specimens was evaluated by seeding them on Vero E6 Cells, a permissive cell line, and monitoring the infection for 7 days. All these procedures have been performed in a Biosecurity level 3 (BLS3) laboratory.
Clinical nasopharyngeal Swab
The clinical specimens were collected from 46 symptomatic coronavirus disease 2019 (COVID-19) individuals at the emergency ward of San Polo hospital of Monfalcone (University Hospital “Giuliano Isontina” – “ASUGI,” Gorizia, Italy) from 28/12/20 to 04/02/21. The patients represented a selection of the 76 SARS-CoV-2-positive patients admitted to the emergency ward (10.2% on a total of 745 patients). They presented fever > 38 °C, cough, and oxygen saturation < 95%. Nasopharyngeal swabs were maintained in universal transport medium UTM (Copan, Brescia Italy) and stored at 4 °C until analyzed.
2019-nCoV kit on the iPonatic instrument
The ORF1ab and N genes were tested with the novel coronavirus (2019-nCoV) kit (S3102E, Sansure Biotech Inc.) on the iPonatic Portable Molecule Workstation (AB Analitica) following the manufacturer’s instruction. Briefly, 50 μl of the samples was loaded in the cassette together with sample release agent (20 μl), PCR mix (20 μl), and enzyme mix (2 μl). After 2 min of thermal extraction, the amplification (3 μl of lysates) was carried out in 40 min (the limit of detection is 200 copies/ml).
STANDARD F COVID-19 Ag FIA
The rapid test STANDARD F COVID-19 Ag FIA (SD BIOSENSOR) was performed accordingly with the manufacturer’s instruction. It is an immunofluorescence lateral flow rapid test, where the samples react with an anti-nucleocapsid SARS-CoV-2 antibody; then, the fluorescence is read with an appropriate analyzer. Briefly, 350 μl of the sample was mixed with the extraction buffer and 4 drops were applied on the specimen well of the test device, and after 30 min, they were read at the STANDARD F200 analyzer. The cutoff index (COI) ≥ 1 was used to interpret the result as positive.
A previously isolated and quantified (in terms of plaque-forming unit-PFU/ml and viral copies/ml) SARS-CoV-2 was used to assess the detection limit of the kit, by performing a serial dilution of the virus (from 1012 to 103 viral copies/ml).
N gene RT-qPCR on 7500 Fast Real-Time PCR instrument
The viral load was quantified through RT-qPCR.
Firstly, 15 μl of the UTM swab was mixed with 45 μl of distilled water and then were subjected to thermolysis (98 °C for 3’ and 4 °C for 5’); then, 3.75 μl was tested with CDC primers and probe (Eurofins, Luxembourg) for the viral gene N (nucleocapsid, 500 nM forward primer GGG AGC CTT GAA TAC ACC AAA A, 500 nM reverse primer TGT AGC ACG ATT GCA GCA TTG, 125 nM probe FAM-AYC ACA TTG GCA CCC GCA ATC CTG-BHQ1), using the Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs, Ipswich, MA, USA), on the 7500 Fast Real-Time PCR instrument (Thermo Fisher Scientific, protocol: 50 °C for 10’, 95 °C for 1’, 40 cycles at 95 °C for 10″, 60° for 30″). A previously quantified nCoV-CDC-Control Plasmid (Eurofins) was used to create the standard curve.
Infectivity
Vero E6 epithelial cell lines from Cercopithecus aethiops kidney (ATCC CRL-1586) were cultured in MEM + 10% fetal bovine serum, 2 mM glutamine, and 100 U/ml penicillin/streptomycin (Euroclone, Pero, Italy).
The day prior to the experiment, cells were seeded at a density of 4 × 105 cells/well in 6 multi-well plates.
One milliliter of each swab was filtered (0.22 μm filter) and seeded on the cells. After 1 h of incubation, the supernatant was harvested, the wells were washed in phosphate-buffered saline, and then, 3 ml of medium (MEM + 2% fetal bovine serum, 2 mM glutamine, and 100 U/ml penicillin/streptomycin, 1 × amphotericin B) were added. After 7 days, the viral load was quantified by using the in-house system for RNA quantification previously described.
The cytopathic effect was assessed with the EVOS XL Core Cell Imaging System (Thermo Fisher Scientific).
Results
Table 1 reports the principal results of our study.
A well concordance was observed in the analysis of the N gene between the 2019-nCoV kit on the iPonatic and the RT-qPCR for the N gene on the 7500 RT-PCR system, with less than 4-cycle thresholds (Ct) of difference. Only for 2 samples, there was a discrepancy of more than 5 Ct. The differences encountered could be ascribed to the different volumes of analysis (both for the extraction and for the PCR).
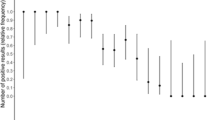
The cutoff index (COI) positivity at the STANDARD F COVID-19 Ag FIA was shown for the specimens presented a Ct value below 31 (at the diagnostic test with iPonatic system), and a great concordance between the iPonatic system and the Ag test was observed when the initial Ct was below 20 (100%) and in the range of 20–25 (92%) and of 25–30 (88%).
Interestingly, the positivity at the antigen test was well correlated to successful infection in the Vero E6 cell line (78% of the cases), but it showed great concordance when the initial Ct was below 20 or above 35 (100%) and in the range 20–25 (83%) (Table 2).
Intriguingly, when the samples were stratified based on the data of collection, some differences were observed. Indeed, the concordance was higher in the first group of patients (from 28/12/20 to 8/01/21) and lower in the second (10/01/21–22/01/21) and the third group (23/01/21–04/02/21) possibly implying some differences related to the SARS-CoV-2 variants circulating in that period (supplementary table 1).
The limit of detection of the antigen assay was also checked with previously isolated SARS-CoV-2: the results are presented in Table 3 and corroborate the findings obtained on the clinical specimens. Indeed, only the dilution with a Ct value < 32 was able to infect the cells and achieve a positive COI at the antigen test.
Discussion
Despite the vaccination campaign has greatly incremented the coverage in the general population, the rise of new variants might impact, depending upon the variant; as an example, the efficacy of the vaccination immunity reaches 5–10% of vaccine effectiveness against Omicron strain after more than 25 weeks from the second dose and 50–60% after 10 weeks from the booster dose with mRNA vaccines BNT162b2-Comirnaty by Pfizer/BioNTech and Fosun Pharma and mRNA-1273—Spikevax, Moderna and National Institute of Allergy and Infectious Diseases [12]. Therefore, the great challenge for the health system is still the early detection of the COVID-19 cases in order to achieve rapid and efficient isolation, especially in health settings where the block of SARS-CoV-2 transmission in the hospital ward is mandatory [1].
In our work, a good concordance was observed between the results from the iPonatic POCT NAAT assay and the in-house quantification system, indicating the effectiveness of the two molecular systems for the detection of the SARS-CoV-2 positive cases, as we can expect. Some differences were observed regarding the concordance when the samples were stratified according to the date range (each group of approximately 10 days), possibly suggesting some influences of the SARS-CoV-2 variants on the efficiency of the tests.
Nevertheless, the more interesting results of our work derived from the antigen test. Indeed, the agreement between the NAAT and the antigen test was particularly evident when the samples presented a low Ct, and therefore, a high viral load, meanwhile, at Ct > 32, the antigen test failed to detect the virus as others previously observed also for assays distributed by other companies [8]. However, the infectivity assessment showed that most of the samples positive at the antigen test were able to establish a persistent infection with virus amplification in vitro on the Vero E6 cells, especially when the initial Ct is lower than 25. This data indicated that the patients from which the swabs were collected presented intact virions and were potential spreaders of the virus. In this work, we decided to test only the subjects positive at the molecular tests since our main aim is related to the assessment of the residual infectious potentiality of the specimens positive to the antigen test.
All the patients analyzed attended the emergency room with symptoms (cough with oxygen saturation < 95% and fever > 38 °C) and were suspected of COVID-19 by the clinicians. Although we are aware that it cannot be excluded their contagiousness based on in vitro assay, performed with the evaluation of the infectivity of the sample on a cell line, it is plausible to think that the patients analyzed in the study presented at the moment of the swab collection weak viral colonization and so the risk of SARS-CoV-2 transmissibility may be very low at that time. These results are in line with those reported by La Scola et al. [13] where the cutoff of 33–34 Ct is the separation between infective and not infective samples.
Generally, the immunofluorescence-based antigen test, like those employed in the current study, showed greater performance in terms of sensibility and specificity with respect to the immunochromatographic colorimetric assays [8], although they are both inferior to the gold standard NAAT. Another type of technology based on microfluidic immunofluorescence, recently developed in the COVID-19 antigen rapid test by LumiraDX claimed a 97.6% positive percent agreement and a 96.6% negative percent agreement within 12 days from symptoms onset, with a 100% positive agreement within 3 days or with Ct < 33 [14].
Independent of the kit employed, antigen rapid tests can be a valid and rapid option to detect the infection; in fact, its use has been evolved and increased during the pandemic. Indeed, the guidelines from the ECDC (lastly updated in October 2021) indicated the antigen test for symptomatic individuals within 5 days from symptoms onset and in a setting with COVID-19 prevalence > 10%. Moreover, the ECDC recommended its employment for testing possible and probable COVID-19 cases presented to the healthcare system to accelerate the COVID-19 case assessment and for contact tracing or outbreak detection in closed settings, as well as in all the cases where the NAAT are not promptly available. In cases of prevalence < 10% positive symptomatic cases should be confirmed by RT-qPCR or by another antigen test from a different producer. For the testing of asymptomatic subjects, the antigen test is recommended when the COVID-19 prevalence was above 10%; otherwise, the RT-qPCR should be preferred. The antigen test could be also employed for early discontinuation of quarantine when resulting negative and the individual is asymptomatic [6, 8].
In Italy, the last guidelines from the Ministry of Health indicated that the antigen test resulted positive does not require a confirmation test for the diagnosis and the isolation of the COVID-19 patients; moreover, a negative test can be employed for the discontinuation of the quarantine. In case of negative test in symptomatic individuals, a second antigenic test or a RT-PCR test should be performed after 2–4 days [15, 16].
The main goal of this type of rapid test would be the mitigation of the huge impact that SARS-CoV-2 diagnosis presents on laboratories in terms of consumables and health workers’ engagement.
Liotti et al. [17] assessed the performance of SENSOR BD STANDARD F COVID-19 Ag FIA on 359 nasopharyngeal specimens demonstrating an agreement with RT-PCR of 98% for the negative cases (i.e., specificity) and of 47% for the positive cases (i.e., sensibility) leading to negative and positive predictive values of 82% and 92.5% respectively. Similar to our findings, below 25 Ct the concordance with NAAT is high (95%); moreover, we observed a concordance of 92% below 30 Ct and a limit of the detection set at 32 Ct, while Liotti et al. reported a low agreement yet up to 25 Ct (42% Ct between 25 and 35, and 21% Ct > 35).
Instead, Porte et al. [18] analyzing 64 nasopharyngeal samples showed specificity of 96.9% and sensitivity of 90.6%, but the assay reached 100% sensitivity when the Ct was under 25.
On the other hand, the presence on the market of POCT molecular tests able to have close to 100% specificity and sensibility would accelerate the diagnostic iter with more reliable data, although the costs are obviously higher with respect to antigen assay, as well as the processivity.
Our results would give some important cues on the potential use of the antigen rapid tests; indeed, our findings showed that most of the swabs positive at the antigen test were able to infect the Vero E6 cells, indicating that probably only these samples present residual infectivity at the moment of collection. In this view, testing the symptomatic individuals with antigen test, it is reasonable to estimate that above all the patients positive at the antigen test are likely to be at higher risk of virus transmission; meanwhile, the individuals negative at the assay could be not able to spread the virus. Indeed, the antigen test employment has been strongly revised and implemented during the pandemic time, being currently used for rapid isolation of symptomatic positive individuals, quarantine discontinuation, and the entry of travelers into countries [8].
Data availability
Not applicable.
Code availability
Not applicable.
References
W.H.O. WHO (2020) Public health surveillance for COVID-19: interim guidance. https://www.who.int/publications-detail-redirect/who-2019-nCoV-surveillanceguidance-2020.8 (Accessed 17 Feb 2021)
E.C. for D.P. and C. ECDC (2020) COVID-19 testing strategies and objectives, European Centre for Disease Prevention and Control. https://www.ecdc.europa.eu/en/publications-data/covid-19-testing-strategies-and-objectives (Accessed 17 Feb 2021)
Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, Emperador D, Takwoingi Y, Cunningham J, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Taylor-Phillips S, Hooft L, Leeflang MM, Spijker R, Van den Bruel A (2020) Cochrane COVID-19 Diagnostic Test Accuracy Group, Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. https://doi.org/10.1002/14651858.CD013705
Serei VD, Cristelli R, Joho K, Salaru G, Kirn T, Carayannopoulous MO, Uprety P (2021) Comparison of Abbott ID NOW COVID-19 rapid molecular assay to Cepheid Xpert Xpress SARS-CoV-2 assay in dry nasal swabs. Diagn Microbiol Infect Dis 99:115208. https://doi.org/10.1016/j.diagmicrobio.2020.115208
Smithgall MC, Dowlatshahi M, Spitalnik SL, Hod EA, Rai AJ (2020) Types of assays for SARS-CoV-2 testing: a review. Lab Med 51:e59–e65. https://doi.org/10.1093/labmed/lmaa039
W.H.O. WHO (2021) Antigen-detection in the diagnosis of SARS-CoV-2 infection. https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays (Accessed 21 Mar 2022)
C. for D.P. and C. CDC (2020) Interim guidance for antigen testing for SARS-CoV-2, Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html#:~:text=Antigen%20tests%20can%20be%20used%20for%20screening%20testing%20in%20high,control%20measures%2C%20thus%20preventing%20transmission (Accessed 17 Feb 2021)
E.C. for D.P. and C. ECDC (2021) Options for the use of rapid antigen tests for COVID-19 in the EU/EEA and the UK - first update, European Centre for Disease Prevention and Control. https://www.ecdc.europa.eu/en/publications-data/options-use-rapid-antigen-tests-covid-19-eueea-first-update (Accessed 21 Mar 2022)
M.P. Romero-Gómez, S. Gómez-Sebastian, E. Cendejas-Bueno, M.D. Montero-Vega, J. Mingorance, J. García-Rodríguez (2020) Ct value is not enough to discriminate patients harbouring infective virus. J Infect S0163445320307209. https://doi.org/10.1016/j.jinf.2020.11.025
Oliveira SC, de Magalhães MTQ, Homan EJ (2020) Immunoinformatic analysis of SARS-CoV-2 nucleocapsid protein and identification of COVID-19 vaccine targets. Front Immunol 11:587615. https://doi.org/10.3389/fimmu.2020.587615
Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, Lynch B, Malapati L, Burke SA, Harcourt J, Tamin A, Thornburg NJ, Villanueva JM, Lindstrom S (2020) US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1654–1665. https://doi.org/10.3201/eid2608.201246
N. Andrews, J. Stowe, F. Kirsebom, S. Toffa, T. Rickeard, E. Gallagher, C. Gower, M. Kall, N. Groves, A.-M. O’Connell, D. Simons, P.B. Blomquist, A. Zaidi, S. Nash, N. Iwani Binti Abdul Aziz, S. Thelwall, G. Dabrera, R. Myers, G. Amirthalingam, S. Gharbia, J.C. Barrett, R. Elson, S.N. Ladhani, N. Ferguson, M. Zambon, C.N.J. Campbell, K. Brown, S. Hopkins, M. Chand, M. Ramsay, J. Lopez Bernal (2022) Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N Engl J Med NEJMoa2119451. https://doi.org/10.1056/NEJMoa2119451
La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, Gautret P, Raoult D (2020) Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 39:1059–1061. https://doi.org/10.1007/s10096-020-03913-9
PK Drain, M. Ampajwala, C. Chappel, AB Gvozden, M. Hoppers, M. Wang, R. Rosen, S. Young, E. Zissman, M. Montano (2020) A rapid, high-sensitivity SARS-CoV-2 nucleocapsid immunoassay to aid diagnosis of acute COVID-19 at the point of care, Infectious Diseases (except HIV/AIDS). https://doi.org/10.1101/2020.12.11.20238410
Ministero della Salute (2021) Aggiornamento sull’uso dei test antigenici e molecolari per la rilevazione di SARS-CoV-2. https://www.trovanorme.salute.gov.it/norme/renderNormsanPdf?anno=2021&codLeg=78828&parte=1%20&serie=null (Accessed 25 Mar 2022)
Ministero della Salute (2021) Aggiornamento sulle misure di quarantena e isolamento in seguito alla diffusione a livello globale della nuova variante VOC SARS-CoV-2 Omicron (B.1.1.529). https://www.trovanorme.salute.gov.it/norme/renderNormsanPdf?anno=2021&codLeg=78828&parte=1%20&serie=null (Accessed 25 Mar 2022)
FM Liotti, G. Menchinelli, E. Lalle, I. Palucci, S. Marchetti, F. Colavita, M. La Sorda, G. Sberna, L. Bordi, M. Sanguinetti, P. Cattani, MR Capobianchi, B. Posteraro (2020) Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clin Microbiol Infect S1198743X20305838. https://doi.org/10.1016/j.cmi.2020.09.030
Porte L, LegarragaP, Iruretagoyena M, Vollrath V, Pizarro G, Munita JM, Araos R, Weitzel T (2020) Rapid SARS-CoV-2 antigen detection by immunofluorescence – a new tool to detect infectivity, Infectious Diseases (except HIV/AIDS). https://doi.org/10.1101/2020.10.04.20206466
Acknowledgements
We are grateful to TCR Tecora—Pollution Check (Cogliane, MB) for providing the BLS3 facility with a microscope. We are grateful to Dr. Giulia Bombi for the English revision of the paper.
Funding
This work was supported by IRCCS Burlo Garofolo /Italian Ministry of Health (RC 15/2017, 03/2020, RC 47/2020).
Author information
Authors and Affiliations
Contributions
L. Z. and L. C. were involved in performing the experiments and writing—original draft. F. F and M. R were involved in the supervision of the experiments conducted in the BLS3 facility, writing—review and editing. S. C. was involved in the conceptualization of the study, writing—review and editing, project management.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Fernando R. Spilki
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zupin, L., Fontana, F., Clemente, L. et al. Comparison between nucleic acid amplification tests, antigen immunofluorescence assay, and in vitro infectivity in SARS-CoV-2 diagnosis. Braz J Microbiol 53, 1271–1277 (2022). https://doi.org/10.1007/s42770-022-00758-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-022-00758-6